Merge from premaster
@ -30,7 +30,7 @@ stages:
|
||||
image: rocker/r-base
|
||||
|
||||
before_script:
|
||||
- apt-get update -qq
|
||||
- apt-get update -qq --allow-releaseinfo-change
|
||||
# install dependencies for packages
|
||||
- apt-get install -y wget locales libxml2-dev libssl-dev libcurl4-openssl-dev zlib1g-dev > /dev/null
|
||||
# recent pandoc
|
||||
@ -116,9 +116,9 @@ coverage:
|
||||
script:
|
||||
- apt-get install --yes git
|
||||
# install missing and outdated packages
|
||||
- Rscript -e 'source(".gitlab-ci.R"); gl_update_pkg_all(repos = "https://cran.rstudio.com", quiet = TRUE, install_pkgdown = TRUE)'
|
||||
- Rscript -e 'source(".gitlab-ci.R"); gl_update_pkg_all(repos = "https://cran.rstudio.com", quiet = TRUE, install_pkgdown = FALSE)'
|
||||
# codecov token is set in https://gitlab.com/msberends/AMR/settings/ci_cd
|
||||
- Rscript -e "cc <- covr::package_coverage(); covr::codecov(coverage = cc, token = '$codecov', exclusions = c('R/atc_online.R', 'R/mo_source.R')); cat('Code coverage:', covr::percent_coverage(cc))"
|
||||
- Rscript -e "cc <- covr::package_coverage(line_exclusions = list('R/atc_online.R', 'R/mo_source.R')); covr::codecov(coverage = cc, token = '$codecov'); cat('Code coverage:', covr::percent_coverage(cc))"
|
||||
coverage: '/Code coverage: \d+\.\d+/'
|
||||
|
||||
pages:
|
||||
|
||||
@ -1,6 +1,6 @@
|
||||
Package: AMR
|
||||
Version: 0.7.1.9008
|
||||
Date: 2019-07-04
|
||||
Version: 0.7.1.9026
|
||||
Date: 2019-08-06
|
||||
Title: Antimicrobial Resistance Analysis
|
||||
Authors@R: c(
|
||||
person(
|
||||
@ -45,6 +45,7 @@ Depends:
|
||||
R (>= 3.1.0)
|
||||
Imports:
|
||||
backports,
|
||||
clean,
|
||||
crayon (>= 1.3.0),
|
||||
data.table (>= 1.9.0),
|
||||
dplyr (>= 0.7.0),
|
||||
@ -59,7 +60,6 @@ Suggests:
|
||||
covr (>= 3.0.1),
|
||||
curl,
|
||||
readxl,
|
||||
rmarkdown,
|
||||
rstudioapi,
|
||||
rvest (>= 0.3.2),
|
||||
testthat (>= 1.0.2),
|
||||
@ -67,7 +67,7 @@ Suggests:
|
||||
VignetteBuilder: knitr
|
||||
URL: https://msberends.gitlab.io/AMR, https://gitlab.com/msberends/AMR
|
||||
BugReports: https://gitlab.com/msberends/AMR/issues
|
||||
License: GPL-2 | file LICENSE
|
||||
License: GPL-2
|
||||
Encoding: UTF-8
|
||||
LazyData: true
|
||||
RoxygenNote: 6.1.1
|
||||
|
||||
339
LICENSE
@ -1,339 +0,0 @@
|
||||
GNU GENERAL PUBLIC LICENSE
|
||||
Version 2, June 1991
|
||||
|
||||
Copyright (C) 1989, 1991 Free Software Foundation, Inc., <http://fsf.org/>
|
||||
51 Franklin Street, Fifth Floor, Boston, MA 02110-1301 USA
|
||||
Everyone is permitted to copy and distribute verbatim copies
|
||||
of this license document, but changing it is not allowed.
|
||||
|
||||
Preamble
|
||||
|
||||
The licenses for most software are designed to take away your
|
||||
freedom to share and change it. By contrast, the GNU General Public
|
||||
License is intended to guarantee your freedom to share and change free
|
||||
software--to make sure the software is free for all its users. This
|
||||
General Public License applies to most of the Free Software
|
||||
Foundation's software and to any other program whose authors commit to
|
||||
using it. (Some other Free Software Foundation software is covered by
|
||||
the GNU Lesser General Public License instead.) You can apply it to
|
||||
your programs, too.
|
||||
|
||||
When we speak of free software, we are referring to freedom, not
|
||||
price. Our General Public Licenses are designed to make sure that you
|
||||
have the freedom to distribute copies of free software (and charge for
|
||||
this service if you wish), that you receive source code or can get it
|
||||
if you want it, that you can change the software or use pieces of it
|
||||
in new free programs; and that you know you can do these things.
|
||||
|
||||
To protect your rights, we need to make restrictions that forbid
|
||||
anyone to deny you these rights or to ask you to surrender the rights.
|
||||
These restrictions translate to certain responsibilities for you if you
|
||||
distribute copies of the software, or if you modify it.
|
||||
|
||||
For example, if you distribute copies of such a program, whether
|
||||
gratis or for a fee, you must give the recipients all the rights that
|
||||
you have. You must make sure that they, too, receive or can get the
|
||||
source code. And you must show them these terms so they know their
|
||||
rights.
|
||||
|
||||
We protect your rights with two steps: (1) copyright the software, and
|
||||
(2) offer you this license which gives you legal permission to copy,
|
||||
distribute and/or modify the software.
|
||||
|
||||
Also, for each author's protection and ours, we want to make certain
|
||||
that everyone understands that there is no warranty for this free
|
||||
software. If the software is modified by someone else and passed on, we
|
||||
want its recipients to know that what they have is not the original, so
|
||||
that any problems introduced by others will not reflect on the original
|
||||
authors' reputations.
|
||||
|
||||
Finally, any free program is threatened constantly by software
|
||||
patents. We wish to avoid the danger that redistributors of a free
|
||||
program will individually obtain patent licenses, in effect making the
|
||||
program proprietary. To prevent this, we have made it clear that any
|
||||
patent must be licensed for everyone's free use or not licensed at all.
|
||||
|
||||
The precise terms and conditions for copying, distribution and
|
||||
modification follow.
|
||||
|
||||
GNU GENERAL PUBLIC LICENSE
|
||||
TERMS AND CONDITIONS FOR COPYING, DISTRIBUTION AND MODIFICATION
|
||||
|
||||
0. This License applies to any program or other work which contains
|
||||
a notice placed by the copyright holder saying it may be distributed
|
||||
under the terms of this General Public License. The "Program", below,
|
||||
refers to any such program or work, and a "work based on the Program"
|
||||
means either the Program or any derivative work under copyright law:
|
||||
that is to say, a work containing the Program or a portion of it,
|
||||
either verbatim or with modifications and/or translated into another
|
||||
language. (Hereinafter, translation is included without limitation in
|
||||
the term "modification".) Each licensee is addressed as "you".
|
||||
|
||||
Activities other than copying, distribution and modification are not
|
||||
covered by this License; they are outside its scope. The act of
|
||||
running the Program is not restricted, and the output from the Program
|
||||
is covered only if its contents constitute a work based on the
|
||||
Program (independent of having been made by running the Program).
|
||||
Whether that is true depends on what the Program does.
|
||||
|
||||
1. You may copy and distribute verbatim copies of the Program's
|
||||
source code as you receive it, in any medium, provided that you
|
||||
conspicuously and appropriately publish on each copy an appropriate
|
||||
copyright notice and disclaimer of warranty; keep intact all the
|
||||
notices that refer to this License and to the absence of any warranty;
|
||||
and give any other recipients of the Program a copy of this License
|
||||
along with the Program.
|
||||
|
||||
You may charge a fee for the physical act of transferring a copy, and
|
||||
you may at your option offer warranty protection in exchange for a fee.
|
||||
|
||||
2. You may modify your copy or copies of the Program or any portion
|
||||
of it, thus forming a work based on the Program, and copy and
|
||||
distribute such modifications or work under the terms of Section 1
|
||||
above, provided that you also meet all of these conditions:
|
||||
|
||||
a) You must cause the modified files to carry prominent notices
|
||||
stating that you changed the files and the date of any change.
|
||||
|
||||
b) You must cause any work that you distribute or publish, that in
|
||||
whole or in part contains or is derived from the Program or any
|
||||
part thereof, to be licensed as a whole at no charge to all third
|
||||
parties under the terms of this License.
|
||||
|
||||
c) If the modified program normally reads commands interactively
|
||||
when run, you must cause it, when started running for such
|
||||
interactive use in the most ordinary way, to print or display an
|
||||
announcement including an appropriate copyright notice and a
|
||||
notice that there is no warranty (or else, saying that you provide
|
||||
a warranty) and that users may redistribute the program under
|
||||
these conditions, and telling the user how to view a copy of this
|
||||
License. (Exception: if the Program itself is interactive but
|
||||
does not normally print such an announcement, your work based on
|
||||
the Program is not required to print an announcement.)
|
||||
|
||||
These requirements apply to the modified work as a whole. If
|
||||
identifiable sections of that work are not derived from the Program,
|
||||
and can be reasonably considered independent and separate works in
|
||||
themselves, then this License, and its terms, do not apply to those
|
||||
sections when you distribute them as separate works. But when you
|
||||
distribute the same sections as part of a whole which is a work based
|
||||
on the Program, the distribution of the whole must be on the terms of
|
||||
this License, whose permissions for other licensees extend to the
|
||||
entire whole, and thus to each and every part regardless of who wrote it.
|
||||
|
||||
Thus, it is not the intent of this section to claim rights or contest
|
||||
your rights to work written entirely by you; rather, the intent is to
|
||||
exercise the right to control the distribution of derivative or
|
||||
collective works based on the Program.
|
||||
|
||||
In addition, mere aggregation of another work not based on the Program
|
||||
with the Program (or with a work based on the Program) on a volume of
|
||||
a storage or distribution medium does not bring the other work under
|
||||
the scope of this License.
|
||||
|
||||
3. You may copy and distribute the Program (or a work based on it,
|
||||
under Section 2) in object code or executable form under the terms of
|
||||
Sections 1 and 2 above provided that you also do one of the following:
|
||||
|
||||
a) Accompany it with the complete corresponding machine-readable
|
||||
source code, which must be distributed under the terms of Sections
|
||||
1 and 2 above on a medium customarily used for software interchange; or,
|
||||
|
||||
b) Accompany it with a written offer, valid for at least three
|
||||
years, to give any third party, for a charge no more than your
|
||||
cost of physically performing source distribution, a complete
|
||||
machine-readable copy of the corresponding source code, to be
|
||||
distributed under the terms of Sections 1 and 2 above on a medium
|
||||
customarily used for software interchange; or,
|
||||
|
||||
c) Accompany it with the information you received as to the offer
|
||||
to distribute corresponding source code. (This alternative is
|
||||
allowed only for noncommercial distribution and only if you
|
||||
received the program in object code or executable form with such
|
||||
an offer, in accord with Subsection b above.)
|
||||
|
||||
The source code for a work means the preferred form of the work for
|
||||
making modifications to it. For an executable work, complete source
|
||||
code means all the source code for all modules it contains, plus any
|
||||
associated interface definition files, plus the scripts used to
|
||||
control compilation and installation of the executable. However, as a
|
||||
special exception, the source code distributed need not include
|
||||
anything that is normally distributed (in either source or binary
|
||||
form) with the major components (compiler, kernel, and so on) of the
|
||||
operating system on which the executable runs, unless that component
|
||||
itself accompanies the executable.
|
||||
|
||||
If distribution of executable or object code is made by offering
|
||||
access to copy from a designated place, then offering equivalent
|
||||
access to copy the source code from the same place counts as
|
||||
distribution of the source code, even though third parties are not
|
||||
compelled to copy the source along with the object code.
|
||||
|
||||
4. You may not copy, modify, sublicense, or distribute the Program
|
||||
except as expressly provided under this License. Any attempt
|
||||
otherwise to copy, modify, sublicense or distribute the Program is
|
||||
void, and will automatically terminate your rights under this License.
|
||||
However, parties who have received copies, or rights, from you under
|
||||
this License will not have their licenses terminated so long as such
|
||||
parties remain in full compliance.
|
||||
|
||||
5. You are not required to accept this License, since you have not
|
||||
signed it. However, nothing else grants you permission to modify or
|
||||
distribute the Program or its derivative works. These actions are
|
||||
prohibited by law if you do not accept this License. Therefore, by
|
||||
modifying or distributing the Program (or any work based on the
|
||||
Program), you indicate your acceptance of this License to do so, and
|
||||
all its terms and conditions for copying, distributing or modifying
|
||||
the Program or works based on it.
|
||||
|
||||
6. Each time you redistribute the Program (or any work based on the
|
||||
Program), the recipient automatically receives a license from the
|
||||
original licensor to copy, distribute or modify the Program subject to
|
||||
these terms and conditions. You may not impose any further
|
||||
restrictions on the recipients' exercise of the rights granted herein.
|
||||
You are not responsible for enforcing compliance by third parties to
|
||||
this License.
|
||||
|
||||
7. If, as a consequence of a court judgment or allegation of patent
|
||||
infringement or for any other reason (not limited to patent issues),
|
||||
conditions are imposed on you (whether by court order, agreement or
|
||||
otherwise) that contradict the conditions of this License, they do not
|
||||
excuse you from the conditions of this License. If you cannot
|
||||
distribute so as to satisfy simultaneously your obligations under this
|
||||
License and any other pertinent obligations, then as a consequence you
|
||||
may not distribute the Program at all. For example, if a patent
|
||||
license would not permit royalty-free redistribution of the Program by
|
||||
all those who receive copies directly or indirectly through you, then
|
||||
the only way you could satisfy both it and this License would be to
|
||||
refrain entirely from distribution of the Program.
|
||||
|
||||
If any portion of this section is held invalid or unenforceable under
|
||||
any particular circumstance, the balance of the section is intended to
|
||||
apply and the section as a whole is intended to apply in other
|
||||
circumstances.
|
||||
|
||||
It is not the purpose of this section to induce you to infringe any
|
||||
patents or other property right claims or to contest validity of any
|
||||
such claims; this section has the sole purpose of protecting the
|
||||
integrity of the free software distribution system, which is
|
||||
implemented by public license practices. Many people have made
|
||||
generous contributions to the wide range of software distributed
|
||||
through that system in reliance on consistent application of that
|
||||
system; it is up to the author/donor to decide if he or she is willing
|
||||
to distribute software through any other system and a licensee cannot
|
||||
impose that choice.
|
||||
|
||||
This section is intended to make thoroughly clear what is believed to
|
||||
be a consequence of the rest of this License.
|
||||
|
||||
8. If the distribution and/or use of the Program is restricted in
|
||||
certain countries either by patents or by copyrighted interfaces, the
|
||||
original copyright holder who places the Program under this License
|
||||
may add an explicit geographical distribution limitation excluding
|
||||
those countries, so that distribution is permitted only in or among
|
||||
countries not thus excluded. In such case, this License incorporates
|
||||
the limitation as if written in the body of this License.
|
||||
|
||||
9. The Free Software Foundation may publish revised and/or new versions
|
||||
of the General Public License from time to time. Such new versions will
|
||||
be similar in spirit to the present version, but may differ in detail to
|
||||
address new problems or concerns.
|
||||
|
||||
Each version is given a distinguishing version number. If the Program
|
||||
specifies a version number of this License which applies to it and "any
|
||||
later version", you have the option of following the terms and conditions
|
||||
either of that version or of any later version published by the Free
|
||||
Software Foundation. If the Program does not specify a version number of
|
||||
this License, you may choose any version ever published by the Free Software
|
||||
Foundation.
|
||||
|
||||
10. If you wish to incorporate parts of the Program into other free
|
||||
programs whose distribution conditions are different, write to the author
|
||||
to ask for permission. For software which is copyrighted by the Free
|
||||
Software Foundation, write to the Free Software Foundation; we sometimes
|
||||
make exceptions for this. Our decision will be guided by the two goals
|
||||
of preserving the free status of all derivatives of our free software and
|
||||
of promoting the sharing and reuse of software generally.
|
||||
|
||||
NO WARRANTY
|
||||
|
||||
11. BECAUSE THE PROGRAM IS LICENSED FREE OF CHARGE, THERE IS NO WARRANTY
|
||||
FOR THE PROGRAM, TO THE EXTENT PERMITTED BY APPLICABLE LAW. EXCEPT WHEN
|
||||
OTHERWISE STATED IN WRITING THE COPYRIGHT HOLDERS AND/OR OTHER PARTIES
|
||||
PROVIDE THE PROGRAM "AS IS" WITHOUT WARRANTY OF ANY KIND, EITHER EXPRESSED
|
||||
OR IMPLIED, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF
|
||||
MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE. THE ENTIRE RISK AS
|
||||
TO THE QUALITY AND PERFORMANCE OF THE PROGRAM IS WITH YOU. SHOULD THE
|
||||
PROGRAM PROVE DEFECTIVE, YOU ASSUME THE COST OF ALL NECESSARY SERVICING,
|
||||
REPAIR OR CORRECTION.
|
||||
|
||||
12. IN NO EVENT UNLESS REQUIRED BY APPLICABLE LAW OR AGREED TO IN WRITING
|
||||
WILL ANY COPYRIGHT HOLDER, OR ANY OTHER PARTY WHO MAY MODIFY AND/OR
|
||||
REDISTRIBUTE THE PROGRAM AS PERMITTED ABOVE, BE LIABLE TO YOU FOR DAMAGES,
|
||||
INCLUDING ANY GENERAL, SPECIAL, INCIDENTAL OR CONSEQUENTIAL DAMAGES ARISING
|
||||
OUT OF THE USE OR INABILITY TO USE THE PROGRAM (INCLUDING BUT NOT LIMITED
|
||||
TO LOSS OF DATA OR DATA BEING RENDERED INACCURATE OR LOSSES SUSTAINED BY
|
||||
YOU OR THIRD PARTIES OR A FAILURE OF THE PROGRAM TO OPERATE WITH ANY OTHER
|
||||
PROGRAMS), EVEN IF SUCH HOLDER OR OTHER PARTY HAS BEEN ADVISED OF THE
|
||||
POSSIBILITY OF SUCH DAMAGES.
|
||||
|
||||
END OF TERMS AND CONDITIONS
|
||||
|
||||
How to Apply These Terms to Your New Programs
|
||||
|
||||
If you develop a new program, and you want it to be of the greatest
|
||||
possible use to the public, the best way to achieve this is to make it
|
||||
free software which everyone can redistribute and change under these terms.
|
||||
|
||||
To do so, attach the following notices to the program. It is safest
|
||||
to attach them to the start of each source file to most effectively
|
||||
convey the exclusion of warranty; and each file should have at least
|
||||
the "copyright" line and a pointer to where the full notice is found.
|
||||
|
||||
{description}
|
||||
Copyright (C) {year} {fullname}
|
||||
|
||||
This program is free software; you can redistribute it and/or modify
|
||||
it under the terms of the GNU General Public License as published by
|
||||
the Free Software Foundation; either version 2 of the License, or
|
||||
(at your option) any later version.
|
||||
|
||||
This program is distributed in the hope that it will be useful,
|
||||
but WITHOUT ANY WARRANTY; without even the implied warranty of
|
||||
MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
|
||||
GNU General Public License for more details.
|
||||
|
||||
You should have received a copy of the GNU General Public License along
|
||||
with this program; if not, write to the Free Software Foundation, Inc.,
|
||||
51 Franklin Street, Fifth Floor, Boston, MA 02110-1301 USA.
|
||||
|
||||
Also add information on how to contact you by electronic and paper mail.
|
||||
|
||||
If the program is interactive, make it output a short notice like this
|
||||
when it starts in an interactive mode:
|
||||
|
||||
Gnomovision version 69, Copyright (C) year name of author
|
||||
Gnomovision comes with ABSOLUTELY NO WARRANTY; for details type `show w'.
|
||||
This is free software, and you are welcome to redistribute it
|
||||
under certain conditions; type `show c' for details.
|
||||
|
||||
The hypothetical commands `show w' and `show c' should show the appropriate
|
||||
parts of the General Public License. Of course, the commands you use may
|
||||
be called something other than `show w' and `show c'; they could even be
|
||||
mouse-clicks or menu items--whatever suits your program.
|
||||
|
||||
You should also get your employer (if you work as a programmer) or your
|
||||
school, if any, to sign a "copyright disclaimer" for the program, if
|
||||
necessary. Here is a sample; alter the names:
|
||||
|
||||
Yoyodyne, Inc., hereby disclaims all copyright interest in the program
|
||||
`Gnomovision' (which makes passes at compilers) written by James Hacker.
|
||||
|
||||
{signature of Ty Coon}, 1 April 1989
|
||||
Ty Coon, President of Vice
|
||||
|
||||
This General Public License does not permit incorporating your program into
|
||||
proprietary programs. If your program is a subroutine library, you may
|
||||
consider it more useful to permit linking proprietary applications with the
|
||||
library. If this is what you want to do, use the GNU Lesser General
|
||||
Public License instead of this License.
|
||||
48
NAMESPACE
@ -1,7 +1,6 @@
|
||||
# Generated by roxygen2: do not edit by hand
|
||||
|
||||
S3method(as.data.frame,ab)
|
||||
S3method(as.data.frame,freq)
|
||||
S3method(as.data.frame,mo)
|
||||
S3method(as.double,mic)
|
||||
S3method(as.integer,mic)
|
||||
@ -10,28 +9,21 @@ S3method(as.rsi,data.frame)
|
||||
S3method(as.rsi,default)
|
||||
S3method(as.rsi,disk)
|
||||
S3method(as.rsi,mic)
|
||||
S3method(as.vector,freq)
|
||||
S3method(as_tibble,freq)
|
||||
S3method(barplot,mic)
|
||||
S3method(barplot,rsi)
|
||||
S3method(boxplot,freq)
|
||||
S3method(diff,freq)
|
||||
S3method(droplevels,mic)
|
||||
S3method(droplevels,rsi)
|
||||
S3method(format,freq)
|
||||
S3method(hist,freq)
|
||||
S3method(freq,mo)
|
||||
S3method(freq,rsi)
|
||||
S3method(kurtosis,data.frame)
|
||||
S3method(kurtosis,default)
|
||||
S3method(kurtosis,matrix)
|
||||
S3method(plot,freq)
|
||||
S3method(plot,mic)
|
||||
S3method(plot,resistance_predict)
|
||||
S3method(plot,rsi)
|
||||
S3method(print,ab)
|
||||
S3method(print,catalogue_of_life_version)
|
||||
S3method(print,disk)
|
||||
S3method(print,freq)
|
||||
S3method(print,frequency_tbl)
|
||||
S3method(print,mic)
|
||||
S3method(print,mo)
|
||||
S3method(print,mo_renamed)
|
||||
@ -39,7 +31,6 @@ S3method(print,mo_uncertainties)
|
||||
S3method(print,rsi)
|
||||
S3method(pull,ab)
|
||||
S3method(pull,mo)
|
||||
S3method(select,freq)
|
||||
S3method(skewness,data.frame)
|
||||
S3method(skewness,default)
|
||||
S3method(skewness,matrix)
|
||||
@ -98,8 +89,6 @@ export(filter_glycopeptides)
|
||||
export(filter_macrolides)
|
||||
export(filter_tetracyclines)
|
||||
export(first_isolate)
|
||||
export(freq)
|
||||
export(frequency_tbl)
|
||||
export(full_join_microorganisms)
|
||||
export(g.test)
|
||||
export(geom_rsi)
|
||||
@ -108,7 +97,6 @@ export(get_mo_source)
|
||||
export(ggplot_rsi)
|
||||
export(ggplot_rsi_predict)
|
||||
export(guess_ab_col)
|
||||
export(header)
|
||||
export(inner_join_microorganisms)
|
||||
export(is.ab)
|
||||
export(is.disk)
|
||||
@ -171,35 +159,26 @@ export(semi_join_microorganisms)
|
||||
export(set_mo_source)
|
||||
export(skewness)
|
||||
export(theme_rsi)
|
||||
export(top_freq)
|
||||
exportMethods(as.data.frame.ab)
|
||||
exportMethods(as.data.frame.freq)
|
||||
exportMethods(as.data.frame.mo)
|
||||
exportMethods(as.double.mic)
|
||||
exportMethods(as.integer.mic)
|
||||
exportMethods(as.numeric.mic)
|
||||
exportMethods(as.vector.freq)
|
||||
exportMethods(as_tibble.freq)
|
||||
exportMethods(barplot.mic)
|
||||
exportMethods(barplot.rsi)
|
||||
exportMethods(boxplot.freq)
|
||||
exportMethods(diff.freq)
|
||||
exportMethods(droplevels.mic)
|
||||
exportMethods(droplevels.rsi)
|
||||
exportMethods(format.freq)
|
||||
exportMethods(hist.freq)
|
||||
exportMethods(freq.mo)
|
||||
exportMethods(freq.rsi)
|
||||
exportMethods(kurtosis)
|
||||
exportMethods(kurtosis.data.frame)
|
||||
exportMethods(kurtosis.default)
|
||||
exportMethods(kurtosis.matrix)
|
||||
exportMethods(plot.freq)
|
||||
exportMethods(plot.mic)
|
||||
exportMethods(plot.rsi)
|
||||
exportMethods(print.ab)
|
||||
exportMethods(print.catalogue_of_life_version)
|
||||
exportMethods(print.disk)
|
||||
exportMethods(print.freq)
|
||||
exportMethods(print.frequency_tbl)
|
||||
exportMethods(print.mic)
|
||||
exportMethods(print.mo)
|
||||
exportMethods(print.mo_renamed)
|
||||
@ -209,7 +188,6 @@ exportMethods(pull.ab)
|
||||
exportMethods(pull.mo)
|
||||
exportMethods(scale_type.ab)
|
||||
exportMethods(scale_type.mo)
|
||||
exportMethods(select.freq)
|
||||
exportMethods(skewness)
|
||||
exportMethods(skewness.data.frame)
|
||||
exportMethods(skewness.default)
|
||||
@ -217,6 +195,8 @@ exportMethods(skewness.matrix)
|
||||
exportMethods(summary.mic)
|
||||
exportMethods(summary.mo)
|
||||
exportMethods(summary.rsi)
|
||||
importFrom(clean,freq)
|
||||
importFrom(clean,top_freq)
|
||||
importFrom(crayon,bgGreen)
|
||||
importFrom(crayon,bgRed)
|
||||
importFrom(crayon,bgYellow)
|
||||
@ -232,7 +212,6 @@ importFrom(crayon,strip_style)
|
||||
importFrom(crayon,underline)
|
||||
importFrom(crayon,white)
|
||||
importFrom(crayon,yellow)
|
||||
importFrom(data.table,address)
|
||||
importFrom(data.table,as.data.table)
|
||||
importFrom(data.table,data.table)
|
||||
importFrom(data.table,setkey)
|
||||
@ -241,9 +220,7 @@ importFrom(dplyr,all_vars)
|
||||
importFrom(dplyr,any_vars)
|
||||
importFrom(dplyr,arrange)
|
||||
importFrom(dplyr,arrange_at)
|
||||
importFrom(dplyr,as_tibble)
|
||||
importFrom(dplyr,between)
|
||||
importFrom(dplyr,bind_cols)
|
||||
importFrom(dplyr,bind_rows)
|
||||
importFrom(dplyr,case_when)
|
||||
importFrom(dplyr,desc)
|
||||
@ -252,7 +229,6 @@ importFrom(dplyr,everything)
|
||||
importFrom(dplyr,filter)
|
||||
importFrom(dplyr,filter_all)
|
||||
importFrom(dplyr,filter_at)
|
||||
importFrom(dplyr,full_join)
|
||||
importFrom(dplyr,funs)
|
||||
importFrom(dplyr,group_by)
|
||||
importFrom(dplyr,group_by_at)
|
||||
@ -275,35 +251,25 @@ importFrom(dplyr,slice)
|
||||
importFrom(dplyr,summarise)
|
||||
importFrom(dplyr,summarise_if)
|
||||
importFrom(dplyr,tibble)
|
||||
importFrom(dplyr,top_n)
|
||||
importFrom(dplyr,transmute)
|
||||
importFrom(dplyr,ungroup)
|
||||
importFrom(dplyr,vars)
|
||||
importFrom(grDevices,boxplot.stats)
|
||||
importFrom(graphics,arrows)
|
||||
importFrom(graphics,axis)
|
||||
importFrom(graphics,barplot)
|
||||
importFrom(graphics,boxplot)
|
||||
importFrom(graphics,hist)
|
||||
importFrom(graphics,par)
|
||||
importFrom(graphics,plot)
|
||||
importFrom(graphics,points)
|
||||
importFrom(graphics,text)
|
||||
importFrom(hms,is.hms)
|
||||
importFrom(knitr,kable)
|
||||
importFrom(microbenchmark,microbenchmark)
|
||||
importFrom(rlang,as_label)
|
||||
importFrom(rlang,enquos)
|
||||
importFrom(rlang,eval_tidy)
|
||||
importFrom(scales,percent)
|
||||
importFrom(stats,complete.cases)
|
||||
importFrom(stats,fivenum)
|
||||
importFrom(stats,glm)
|
||||
importFrom(stats,lm)
|
||||
importFrom(stats,mad)
|
||||
importFrom(stats,pchisq)
|
||||
importFrom(stats,predict)
|
||||
importFrom(stats,sd)
|
||||
importFrom(utils,browseURL)
|
||||
importFrom(utils,browseVignettes)
|
||||
importFrom(utils,installed.packages)
|
||||
importFrom(utils,menu)
|
||||
|
||||
17
NEWS.md
@ -1,4 +1,7 @@
|
||||
# AMR 0.7.1.9008
|
||||
# AMR 0.7.1.9026
|
||||
|
||||
### Breaking
|
||||
* Function `freq()` has moved to a new package, [`clean`](https://github.com/msberends/clean) ([CRAN link](https://cran.r-project.org/package=clean)). Creating frequency tables is actually not the scope of this package (never was) and this function has matured a lot over the last two years. Therefore, a new package was created for data cleaning and checking and it perfectly fits the `freq()` function. The [`clean`](https://github.com/msberends/clean) package is available on CRAN and will be installed automatically when updating the `AMR` package, that now imports it. In a later stage, the `skewness()` and `kurtosis()` functions will be moved to the `clean` package too.
|
||||
|
||||
### New
|
||||
* Additional way to calculate co-resistance, i.e. when using multiple antibiotics as input for `portion_*` functions or `count_*` functions. This can be used to determine the empiric susceptibily of a combination therapy. A new parameter `only_all_tested` (**which defaults to `FALSE`**) replaces the old `also_single_tested` and can be used to select one of the two methods to count isolates and calculate portions. The difference can be seen in this example table (which is also on the `portion` and `count` help pages), where the %SI is being determined:
|
||||
@ -32,15 +35,21 @@
|
||||
Since this is a major change, usage of the old `also_single_tested` will throw an informative error that it has been replaced by `only_all_tested`.
|
||||
|
||||
### Changed
|
||||
* Fixed a bug in `eucast_rules()` that caused an error when the input was a specific kind of `tibble`
|
||||
* Removed class `atc` - using `as.atc()` is now deprecated in favour of `ab_atc()` and this will return a character, not the `atc` class anymore
|
||||
* Removed deprecated functions `abname()`, `ab_official()`, `atc_name()`, `atc_official()`, `atc_property()`, `atc_tradenames()`, `atc_trivial_nl()`
|
||||
* Fix and speed improvement for `mo_shortname()`
|
||||
* Fix for `as.mo()` where misspelled input would not be understood
|
||||
* Algorithm improvements for `as.mo()`:
|
||||
* Some misspelled input were not understood
|
||||
* These new trivial names known to the field are now understood: meningococcus, gonococcus, pneumococcus
|
||||
* Added support for unknown yeasts and fungi
|
||||
* Added the newest taxonomic data from the IJSEM journal (now up to date until August 2019)
|
||||
* Fix for using `mo_*` functions where the coercion uncertainties and failures would not be available through `mo_uncertainties()` and `mo_failures()` anymore
|
||||
* Deprecated the `country` parameter of `mdro()` in favour of the already existing `guideline` parameter to support multiple guidelines within one country
|
||||
* Fix for frequency tables when creating one directly on a group (using `group_by()`)
|
||||
* The name of `RIF` is now Rifampicin instead of Rifampin
|
||||
* The `name` of `RIF` is now Rifampicin instead of Rifampin
|
||||
* The `antibiotics` data set is now sorted by name
|
||||
* Using verbose mode with `eucast_rules(..., verbose = TRUE)` returns more informative and readable output
|
||||
* Speed improvement for `guess_ab_col()` which is now 30 times faster for antibiotic abbreviations
|
||||
|
||||
# AMR 0.7.1
|
||||
|
||||
|
||||
@ -90,6 +90,7 @@ NULL
|
||||
#' @export
|
||||
#' @examples
|
||||
#' library(dplyr)
|
||||
#' library(clean)
|
||||
#' microorganisms %>% freq(kingdom)
|
||||
#' microorganisms %>% group_by(kingdom) %>% freq(phylum, nmax = NULL)
|
||||
catalogue_of_life_version <- function() {
|
||||
|
||||
10
R/data.R
@ -55,7 +55,7 @@
|
||||
#'
|
||||
#' A data set containing the microbial taxonomy of six kingdoms from the Catalogue of Life. MO codes can be looked up using \code{\link{as.mo}}.
|
||||
#' @inheritSection catalogue_of_life Catalogue of Life
|
||||
#' @format A \code{\link{data.frame}} with 67,906 observations and 16 variables:
|
||||
#' @format A \code{\link{data.frame}} with 68,260 observations and 16 variables:
|
||||
#' \describe{
|
||||
#' \item{\code{mo}}{ID of microorganism as used by this package}
|
||||
#' \item{\code{col_id}}{Catalogue of Life ID}
|
||||
@ -72,8 +72,8 @@
|
||||
#' \item{9 entries of \emph{Streptococcus} (beta haemolytic groups A, B, C, D, F, G, H, K and unspecified)}
|
||||
#' \item{2 entries of \emph{Staphylococcus} (coagulase-negative [CoNS] and coagulase-positive [CoPS])}
|
||||
#' \item{3 entries of Trichomonas (Trichomonas vaginalis, and its family and genus)}
|
||||
#' \item{3 other 'undefined' entries (unknown, unknown Gram negatives and unknown Gram positives)}
|
||||
#' \item{8,830 species from the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen) that are not in the Catalogue of Life}
|
||||
#' \item{5 other 'undefined' entries (unknown, unknown Gram negatives, unknown Gram positives, unknown yeast and unknown fungus)}
|
||||
#' \item{8,970 species from the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen) that are not in the Catalogue of Life}
|
||||
#' }
|
||||
#' @section About the records from DSMZ (see source):
|
||||
#' Names of prokaryotes are defined as being validly published by the International Code of Nomenclature of Bacteria. Validly published are all names which are included in the Approved Lists of Bacterial Names and the names subsequently published in the International Journal of Systematic Bacteriology (IJSB) and, from January 2000, in the International Journal of Systematic and Evolutionary Microbiology (IJSEM) as original articles or in the validation lists.
|
||||
@ -91,14 +91,14 @@ catalogue_of_life <- list(
|
||||
version = "Catalogue of Life: {year} Annual Checklist",
|
||||
url_CoL = "http://www.catalogueoflife.org/annual-checklist/{year}/",
|
||||
url_DSMZ = "https://www.dsmz.de/microorganisms/pnu/bacterial_nomenclature_info_mm.php",
|
||||
yearmonth_DSMZ = "February 2019"
|
||||
yearmonth_DSMZ = "August 2019"
|
||||
)
|
||||
|
||||
#' Data set with previously accepted taxonomic names
|
||||
#'
|
||||
#' A data set containing old (previously valid or accepted) taxonomic names according to the Catalogue of Life. This data set is used internally by \code{\link{as.mo}}.
|
||||
#' @inheritSection catalogue_of_life Catalogue of Life
|
||||
#' @format A \code{\link{data.frame}} with 21,342 observations and 4 variables:
|
||||
#' @format A \code{\link{data.frame}} with 21,743 observations and 4 variables:
|
||||
#' \describe{
|
||||
#' \item{\code{col_id}}{Catalogue of Life ID that was originally given}
|
||||
#' \item{\code{col_id_new}}{New Catalogue of Life ID that responds to an entry in the \code{\link{microorganisms}} data set}
|
||||
|
||||
@ -29,7 +29,7 @@ EUCAST_VERSION_EXPERT_RULES <- "3.1, 2016"
|
||||
#' @param x data with antibiotic columns, like e.g. \code{AMX} and \code{AMC}
|
||||
#' @param info print progress
|
||||
#' @param rules a character vector that specifies which rules should be applied - one or more of \code{c("breakpoints", "expert", "other", "all")}
|
||||
#' @param verbose a logical to indicate whether extensive info should be returned as a \code{data.frame} with info about which rows and columns are effected. It runs all EUCAST rules, but will not be applied to an output - only an informative \code{data.frame} with changes will be returned as output.
|
||||
#' @param verbose a logical to turn Verbose mode on and off (default is off). In Verbose mode, the function does not apply rules to the data, but instead returns a \code{data.frame} with extensive info about which rows and columns would be effected and in which way.
|
||||
#' @param ... column name of an antibiotic, see section Antibiotics
|
||||
#' @inheritParams first_isolate
|
||||
#' @details
|
||||
@ -119,7 +119,8 @@ EUCAST_VERSION_EXPERT_RULES <- "3.1, 2016"
|
||||
#' @rdname eucast_rules
|
||||
#' @export
|
||||
#' @importFrom dplyr %>% select pull mutate_at vars group_by summarise n
|
||||
#' @importFrom crayon bold bgGreen bgYellow bgRed black green blue italic strip_style white
|
||||
#' @importFrom crayon bold bgGreen bgYellow bgRed black green blue italic strip_style white red
|
||||
#' @importFrom utils menu
|
||||
#' @return The input of \code{x}, possibly with edited values of antibiotics. Or, if \code{verbose = TRUE}, a \code{data.frame} with all original and new values of the affected bug-drug combinations.
|
||||
#' @source
|
||||
#' \itemize{
|
||||
@ -139,8 +140,6 @@ EUCAST_VERSION_EXPERT_RULES <- "3.1, 2016"
|
||||
#' }
|
||||
#' @inheritSection AMR Read more on our website!
|
||||
#' @examples
|
||||
#' a <- eucast_rules(septic_patients)
|
||||
#'
|
||||
#' a <- data.frame(mo = c("Staphylococcus aureus",
|
||||
#' "Enterococcus faecalis",
|
||||
#' "Escherichia coli",
|
||||
@ -176,7 +175,7 @@ EUCAST_VERSION_EXPERT_RULES <- "3.1, 2016"
|
||||
#' # 5 Pseudomonas aeruginosa R R - - R R R
|
||||
#'
|
||||
#'
|
||||
#' # do not apply EUCAST rules, but rather get a a data.frame
|
||||
#' # do not apply EUCAST rules, but rather get a data.frame
|
||||
#' # with 18 rows, containing all details about the transformations:
|
||||
#' c <- eucast_rules(a, verbose = TRUE)
|
||||
eucast_rules <- function(x,
|
||||
@ -185,9 +184,21 @@ eucast_rules <- function(x,
|
||||
rules = c("breakpoints", "expert", "other", "all"),
|
||||
verbose = FALSE,
|
||||
...) {
|
||||
|
||||
x <- x
|
||||
|
||||
|
||||
if (verbose == TRUE & interactive()) {
|
||||
txt <- paste0("WARNING: In Verbose mode, the eucast_rules() function does not apply rules to the data, but instead returns a data set in logbook form: with extensive info about which rows and columns would be effected and in which way.",
|
||||
"\n\nThis may overwrite your existing data if you use e.g.:",
|
||||
"\ndata <- eucast_rules(data, verbose = TRUE)\n\nDo you want to continue?")
|
||||
if ("rstudioapi" %in% rownames(installed.packages())) {
|
||||
q_continue <- rstudioapi::showQuestion("Using verbose = TRUE with eucast_rules()", txt)
|
||||
} else {
|
||||
q_continue <- menu(choices = c("OK", "Cancel"), graphics = TRUE, title = txt)
|
||||
}
|
||||
if (q_continue %in% c(FALSE, 2)) {
|
||||
return(invisible())
|
||||
}
|
||||
}
|
||||
|
||||
if (!is.data.frame(x)) {
|
||||
stop("`x` must be a data frame.", call. = FALSE)
|
||||
}
|
||||
@ -385,7 +396,6 @@ eucast_rules <- function(x,
|
||||
cols <- unique(cols[!is.na(cols) & !is.null(cols)])
|
||||
if (length(rows) > 0 & length(cols) > 0) {
|
||||
before_df <- x_original
|
||||
before <- as.character(unlist(as.list(x_original[rows, cols])))
|
||||
|
||||
tryCatch(
|
||||
# insert into original table
|
||||
@ -406,9 +416,7 @@ eucast_rules <- function(x,
|
||||
|
||||
x[rows, cols] <<- x_original[rows, cols]
|
||||
|
||||
after <- as.character(unlist(as.list(x_original[rows, cols])))
|
||||
|
||||
# before_df might not be a data.frame, but a tibble of data.table instead
|
||||
# before_df might not be a data.frame, but a tibble or data.table instead
|
||||
old <- as.data.frame(before_df, stringsAsFactors = FALSE)[rows,]
|
||||
no_of_changes_this_run <- 0
|
||||
for (i in 1:length(cols)) {
|
||||
@ -423,13 +431,14 @@ eucast_rules <- function(x,
|
||||
stringsAsFactors = FALSE)
|
||||
colnames(verbose_new) <- c("row", "col", "mo_fullname", "old", "new", "rule", "rule_group", "rule_name")
|
||||
verbose_new <- verbose_new %>% filter(old != new | is.na(old))
|
||||
# save changes to data set 'verbose_info'
|
||||
verbose_info <<- rbind(verbose_info, verbose_new)
|
||||
no_of_changes_this_run <- no_of_changes_this_run + nrow(verbose_new)
|
||||
}
|
||||
# return number of (new) changes
|
||||
# after the applied changes: return number of (new) changes
|
||||
return(no_of_changes_this_run)
|
||||
}
|
||||
# return number of (new) changes: none.
|
||||
# no changes were applied: return number of (new) changes: none.
|
||||
return(0)
|
||||
}
|
||||
|
||||
@ -498,6 +507,15 @@ eucast_rules <- function(x,
|
||||
}
|
||||
y[y != "" & y %in% colnames(df)]
|
||||
}
|
||||
get_antibiotic_names <- function(x) {
|
||||
x %>%
|
||||
strsplit(",") %>%
|
||||
unlist() %>%
|
||||
trimws() %>%
|
||||
sapply(function(x) if(x %in% AMR::antibiotics$ab) ab_name(x, language = NULL, tolower = TRUE) else x) %>%
|
||||
sort() %>%
|
||||
paste(collapse = ", ")
|
||||
}
|
||||
|
||||
eucast_rules_df <- eucast_rules_file # internal data file
|
||||
no_of_changes <- 0
|
||||
@ -510,10 +528,11 @@ eucast_rules <- function(x,
|
||||
rule_group_current <- eucast_rules_df[i, "reference.rule_group"]
|
||||
rule_group_next <- eucast_rules_df[min(nrow(eucast_rules_df), i + 1), "reference.rule_group"]
|
||||
if (is.na(eucast_rules_df[i, 4])) {
|
||||
rule_text <- paste("always:", eucast_rules_df[i, 6], "=", eucast_rules_df[i, 7])
|
||||
rule_text <- paste0("always report as '", eucast_rules_df[i, 7], "': ", get_antibiotic_names(eucast_rules_df[i, 6]))
|
||||
} else {
|
||||
rule_text <- paste("if", eucast_rules_df[i, 4], "=", eucast_rules_df[i, 5],
|
||||
"then", eucast_rules_df[i, 6], "=", eucast_rules_df[i, 7])
|
||||
rule_text <- paste0("report as '", eucast_rules_df[i, 7], "' when ",
|
||||
get_antibiotic_names(eucast_rules_df[i, 4]), " is '", eucast_rules_df[i, 5], "': ",
|
||||
get_antibiotic_names(eucast_rules_df[i, 6]))
|
||||
}
|
||||
if (i == 1) {
|
||||
rule_previous <- ""
|
||||
@ -736,7 +755,9 @@ eucast_rules <- function(x,
|
||||
cat(paste0(silver(strrep("-", options()$width - 1)), "\n"))
|
||||
|
||||
if (verbose == FALSE & nrow(verbose_info) > 0) {
|
||||
cat(paste("\nUse", bold("verbose = TRUE"), "to get a data.frame with all specified edits instead.\n"))
|
||||
cat(paste("\nUse", bold("verbose = TRUE"), "(on your original data) to get a data.frame with all specified edits instead.\n"))
|
||||
} else if (verbose == TRUE) {
|
||||
cat(paste(red("\nUsed 'Verbose mode' (verbose = TRUE)."), "This returns a data.frame with all specified edits.\nUse", bold("verbose = FALSE"), "to apply the rules on your data.\n"))
|
||||
}
|
||||
}
|
||||
|
||||
|
||||
@ -70,7 +70,7 @@
|
||||
#' @keywords isolate isolates first
|
||||
#' @seealso \code{\link{key_antibiotics}}
|
||||
#' @export
|
||||
#' @importFrom dplyr arrange_at lag between row_number filter mutate arrange pull
|
||||
#' @importFrom dplyr arrange_at lag between row_number filter mutate arrange pull ungroup
|
||||
#' @importFrom crayon blue bold silver
|
||||
#' @return Logical vector
|
||||
#' @source Methodology of this function is based on: \strong{M39 Analysis and Presentation of Cumulative Antimicrobial Susceptibility Test Data, 4th Edition}, 2014, \emph{Clinical and Laboratory Standards Institute (CLSI)}. \url{https://clsi.org/standards/products/microbiology/documents/m39/}.
|
||||
|
||||
@ -67,7 +67,10 @@ get_locale <- function() {
|
||||
}
|
||||
|
||||
lang <- Sys.getlocale("LC_COLLATE")
|
||||
# grepl with case = FALSE is faster than like
|
||||
|
||||
# Check the locale settings for a start with one of these languages:
|
||||
|
||||
# grepl() with ignore.case = FALSE is faster than %like%
|
||||
if (grepl("^(English|en_|EN_)", lang, ignore.case = FALSE)) {
|
||||
# as first option to optimise speed
|
||||
"en"
|
||||
|
||||
@ -79,6 +79,13 @@ guess_ab_col <- function(x = NULL, search_string = NULL, verbose = FALSE) {
|
||||
search_string.ab <- suppressWarnings(as.ab(search_string))
|
||||
if (search_string.ab %in% colnames(x)) {
|
||||
ab_result <- colnames(x)[colnames(x) == search_string.ab][1L]
|
||||
|
||||
} else if (any(tolower(colnames(x)) %in% tolower(unlist(ab_property(search_string.ab, "abbreviations"))))) {
|
||||
ab_result <- colnames(x)[tolower(colnames(x)) %in% tolower(unlist(ab_property(search_string.ab, "abbreviations")))][1L]
|
||||
|
||||
# } else if (any(tolower(colnames(x)) %in% tolower(ab_tradenames(search_string.ab)))) {
|
||||
# ab_result <- colnames(x)[tolower(colnames(x)) %in% tolower(ab_tradenames(search_string.ab))][1L]
|
||||
|
||||
} else {
|
||||
# sort colnames on length - longest first
|
||||
cols <- colnames(x[, x %>% colnames() %>% nchar() %>% order() %>% rev()])
|
||||
|
||||
7
R/like.R
@ -48,6 +48,7 @@
|
||||
#'
|
||||
#' # get frequencies of bacteria whose name start with 'Ent' or 'ent'
|
||||
#' library(dplyr)
|
||||
#' library(clean)
|
||||
#' septic_patients %>%
|
||||
#' left_join_microorganisms() %>%
|
||||
#' filter(genus %like% '^ent') %>%
|
||||
@ -75,7 +76,11 @@ like <- function(x, pattern) {
|
||||
if (is.factor(x)) {
|
||||
as.integer(x) %in% base::grep(pattern, levels(x), ignore.case = TRUE)
|
||||
} else {
|
||||
base::grepl(pattern, x, ignore.case = TRUE)
|
||||
tryCatch(base::grepl(pattern, x, ignore.case = TRUE),
|
||||
error = function(e) ifelse(test = grepl("Invalid regexp", e$message),
|
||||
# try with perl = TRUE:
|
||||
yes = return(base::grepl(pattern, x, ignore.case = TRUE, perl = TRUE)),
|
||||
no = stop(e$message)))
|
||||
}
|
||||
}
|
||||
|
||||
|
||||
4
R/mdro.R
@ -496,8 +496,8 @@ mdro <- function(x,
|
||||
|
||||
#' @rdname mdro
|
||||
#' @export
|
||||
brmo <- function(..., guideline = "BRMO") {
|
||||
mdro(..., guideline = "BRMO")
|
||||
brmo <- function(x, guideline = "BRMO", ...) {
|
||||
mdro(x, guideline = "BRMO", ...)
|
||||
}
|
||||
|
||||
#' @rdname mdro
|
||||
|
||||
2
R/mic.R
@ -51,6 +51,8 @@
|
||||
#'
|
||||
#' plot(mic_data)
|
||||
#' barplot(mic_data)
|
||||
#'
|
||||
#' library(clean)
|
||||
#' freq(mic_data)
|
||||
as.mic <- function(x, na.rm = FALSE) {
|
||||
if (is.mic(x)) {
|
||||
|
||||
28
R/misc.R
@ -71,18 +71,20 @@ size_humanreadable <- function(bytes, decimals = 1) {
|
||||
out
|
||||
}
|
||||
|
||||
percent_scales <- scales::percent
|
||||
percent_clean <- clean:::percent
|
||||
# No export, no Rd
|
||||
# based on scales::percent
|
||||
percent <- function(x, round = 1, force_zero = FALSE, decimal.mark = getOption("OutDec"), ...) {
|
||||
x <- percent_scales(x = as.double(x),
|
||||
accuracy = 1 / 10 ^ round,
|
||||
decimal.mark = decimal.mark,
|
||||
...)
|
||||
if (force_zero == FALSE) {
|
||||
x <- gsub("([.]%|%%)", "%", paste0(gsub("0+%$", "", x), "%"))
|
||||
percent <- function(x, round = 1, force_zero = FALSE, decimal.mark = getOption("OutDec"), big.mark = ",", ...) {
|
||||
if (decimal.mark == big.mark) {
|
||||
if (decimal.mark == ",") {
|
||||
big.mark <- "."
|
||||
} else if (decimal.mark == ".") {
|
||||
big.mark <- ","
|
||||
} else {
|
||||
big.mark <- " "
|
||||
}
|
||||
}
|
||||
x
|
||||
x <- percent_clean(x = x, round = round, force_zero = force_zero,
|
||||
decimal.mark = decimal.mark, big.mark = big.mark, ...)
|
||||
}
|
||||
|
||||
#' @importFrom crayon blue bold red
|
||||
@ -97,8 +99,10 @@ search_type_in_df <- function(x, type) {
|
||||
if (type == "mo") {
|
||||
if ("mo" %in% lapply(x, class)) {
|
||||
found <- colnames(x)[lapply(x, class) == "mo"][1]
|
||||
} else if (any(colnames(x) %like% "^(mo|microorganism|organism|bacteria)s?$")) {
|
||||
found <- colnames(x)[colnames(x) %like% "^(mo|microorganism|organism|bacteria)s?$"][1]
|
||||
} else if (any(colnames(x) %like% "^(mo|microorganism|organism|bacteria|bacterie)s?$")) {
|
||||
found <- colnames(x)[colnames(x) %like% "^(mo|microorganism|organism|bacteria|bacterie)s?$"][1]
|
||||
} else if (any(colnames(x) %like% "^(microorganism|organism|bacteria|bacterie)")) {
|
||||
found <- colnames(x)[colnames(x) %like% "^(microorganism|organism|bacteria|bacterie)"][1]
|
||||
} else if (any(colnames(x) %like% "species")) {
|
||||
found <- colnames(x)[colnames(x) %like% "species"][1]
|
||||
}
|
||||
|
||||
128
R/mo.R
@ -472,7 +472,9 @@ exec_as.mo <- function(x,
|
||||
x_backup_without_spp <- x
|
||||
x_species <- paste(x, "species")
|
||||
# translate to English for supported languages of mo_property
|
||||
x <- gsub("(Gruppe|gruppe|groep|grupo|gruppo|groupe)", "group", x, ignore.case = TRUE)
|
||||
x <- gsub("(gruppe|groep|grupo|gruppo|groupe)", "group", x, ignore.case = TRUE)
|
||||
x <- gsub("(hefe|gist|gisten|levadura|lievito|fermento|levure)[a-z]*", "yeast", x, ignore.case = TRUE)
|
||||
x <- gsub("(schimmels?|mofo|molde|stampo|moisissure)[a-z]*", "fungus", x, ignore.case = TRUE)
|
||||
# remove non-text in case of "E. coli" except dots and spaces
|
||||
x <- gsub("[^.a-zA-Z0-9/ \\-]+", "", x)
|
||||
# replace minus by a space
|
||||
@ -483,7 +485,7 @@ exec_as.mo <- function(x,
|
||||
x <- gsub("(alpha|beta|gamma).?ha?emoly", "\\1-haemoly", x)
|
||||
# remove genus as first word
|
||||
x <- gsub("^Genus ", "", x)
|
||||
# allow characters that resemble others
|
||||
# allow characters that resemble others ----
|
||||
if (initial_search == FALSE) {
|
||||
x <- tolower(x)
|
||||
x <- gsub("[iy]+", "[iy]+", x)
|
||||
@ -493,15 +495,17 @@ exec_as.mo <- function(x,
|
||||
x <- gsub("a+", "a+", x)
|
||||
x <- gsub("u+", "u+", x)
|
||||
# allow any ending of -um, -us, -ium, -icum, -ius, -icus, -ica and -a (needs perl for the negative backward lookup):
|
||||
x <- gsub("(u\\+\\(c\\|k\\|q\\|qu\\+\\|s\\|z\\|x\\|ks\\)\\+)(?![a-z[])",
|
||||
x <- gsub("(u\\+\\(c\\|k\\|q\\|qu\\+\\|s\\|z\\|x\\|ks\\)\\+)(?![a-z])",
|
||||
"(u[s|m]|[iy][ck]?u[ms]|[iy]?[ck]?a)", x, ignore.case = TRUE, perl = TRUE)
|
||||
x <- gsub("(\\[iy\\]\\+\\(c\\|k\\|q\\|qu\\+\\|s\\|z\\|x\\|ks\\)\\+a\\+)(?![a-z[])",
|
||||
x <- gsub("(\\[iy\\]\\+\\(c\\|k\\|q\\|qu\\+\\|s\\|z\\|x\\|ks\\)\\+a\\+)(?![a-z])",
|
||||
"(u[s|m]|[iy][ck]?u[ms]|[iy]?[ck]?a)", x, ignore.case = TRUE, perl = TRUE)
|
||||
x <- gsub("(\\[iy\\]\\+u\\+m)(?![a-z[])",
|
||||
x <- gsub("(\\[iy\\]\\+u\\+m)(?![a-z])",
|
||||
"(u[s|m]|[iy][ck]?u[ms]|[iy]?[ck]?a)", x, ignore.case = TRUE, perl = TRUE)
|
||||
x <- gsub("e+", "e+", x, ignore.case = TRUE)
|
||||
x <- gsub("o+", "o+", x, ignore.case = TRUE)
|
||||
x <- gsub("(.)\\1+", "\\1+", x)
|
||||
# allow ending in -en or -us
|
||||
x <- gsub("e\\+n(?![a-z[])", "(e+n|u+(c|k|q|qu|s|z|x|ks)+)", x, ignore.case = TRUE, perl = TRUE)
|
||||
}
|
||||
x <- strip_whitespace(x)
|
||||
|
||||
@ -519,7 +523,7 @@ exec_as.mo <- function(x,
|
||||
x_withspaces_end_only <- paste0(x_withspaces, '$')
|
||||
x_withspaces_start_end <- paste0('^', x_withspaces, '$')
|
||||
|
||||
if (debug == TRUE) {
|
||||
if (isTRUE(debug)) {
|
||||
cat(paste0('x "', x, '"\n'))
|
||||
cat(paste0('x_species "', x_species, '"\n'))
|
||||
cat(paste0('x_withspaces_start_only "', x_withspaces_start_only, '"\n'))
|
||||
@ -725,6 +729,14 @@ exec_as.mo <- function(x,
|
||||
}
|
||||
next
|
||||
}
|
||||
if (x_backup_without_spp[i] %like% 'haemoly.*strept') {
|
||||
# Haemolytic streptococci in different languages
|
||||
x[i] <- microorganismsDT[mo == 'B_STRPT_HAE', ..property][[1]][1L]
|
||||
if (initial_search == TRUE) {
|
||||
set_mo_history(x_backup[i], get_mo_code(x[i], property), 0, force = force_mo_history)
|
||||
}
|
||||
next
|
||||
}
|
||||
# CoNS/CoPS in different languages (support for German, Dutch, Spanish, Portuguese) ----
|
||||
if (x_backup_without_spp[i] %like% '[ck]oagulas[ea] negatie?[vf]'
|
||||
| x_trimmed[i] %like% '[ck]oagulas[ea] negatie?[vf]'
|
||||
@ -787,6 +799,32 @@ exec_as.mo <- function(x,
|
||||
}
|
||||
next
|
||||
}
|
||||
|
||||
# trivial names known to the field:
|
||||
if ("meningococcus" %like% x_trimmed[i]) {
|
||||
# coerce S. coagulase positive
|
||||
x[i] <- microorganismsDT[mo == 'B_NESSR_MEN', ..property][[1]][1L]
|
||||
if (initial_search == TRUE) {
|
||||
set_mo_history(x_backup[i], get_mo_code(x[i], property), 0, force = force_mo_history)
|
||||
}
|
||||
next
|
||||
}
|
||||
if ("gonococcus" %like% x_trimmed[i]) {
|
||||
# coerce S. coagulase positive
|
||||
x[i] <- microorganismsDT[mo == 'B_NESSR_GON', ..property][[1]][1L]
|
||||
if (initial_search == TRUE) {
|
||||
set_mo_history(x_backup[i], get_mo_code(x[i], property), 0, force = force_mo_history)
|
||||
}
|
||||
next
|
||||
}
|
||||
if ("pneumococcus" %like% x_trimmed[i]) {
|
||||
# coerce S. coagulase positive
|
||||
x[i] <- microorganismsDT[mo == 'B_STRPT_PNE', ..property][[1]][1L]
|
||||
if (initial_search == TRUE) {
|
||||
set_mo_history(x_backup[i], get_mo_code(x[i], property), 0, force = force_mo_history)
|
||||
}
|
||||
next
|
||||
}
|
||||
}
|
||||
|
||||
# FIRST TRY FULLNAMES AND CODES ----
|
||||
@ -1006,6 +1044,9 @@ exec_as.mo <- function(x,
|
||||
|
||||
if (uncertainty_level >= 1) {
|
||||
# (1) look again for old taxonomic names, now for G. species ----
|
||||
if (isTRUE(debug)) {
|
||||
cat("\n[UNCERTAINLY LEVEL 1] (1) look again for old taxonomic names, now for G. species\n")
|
||||
}
|
||||
found <- microorganisms.oldDT[fullname %like% c.x_withspaces_start_end
|
||||
| fullname %like% d.x_withspaces_start_only]
|
||||
if (NROW(found) > 0 & nchar(g.x_backup_without_spp) >= 6) {
|
||||
@ -1035,7 +1076,10 @@ exec_as.mo <- function(x,
|
||||
|
||||
# (2) Try with misspelled input ----
|
||||
# just rerun with initial_search = FALSE will used the extensive regex part above
|
||||
found <- suppressMessages(suppressWarnings(exec_as.mo(a.x_backup, initial_search = FALSE, allow_uncertain = FALSE)))
|
||||
if (isTRUE(debug)) {
|
||||
cat("\n[UNCERTAINLY LEVEL 1] (2) Try with misspelled input\n")
|
||||
}
|
||||
found <- suppressMessages(suppressWarnings(exec_as.mo(a.x_backup, initial_search = FALSE, allow_uncertain = FALSE, debug = debug)))
|
||||
if (!empty_result(found)) {
|
||||
found_result <- found
|
||||
found <- microorganismsDT[mo == found, ..property][[1]]
|
||||
@ -1054,6 +1098,9 @@ exec_as.mo <- function(x,
|
||||
if (uncertainty_level >= 2) {
|
||||
|
||||
# (3) look for genus only, part of name ----
|
||||
if (isTRUE(debug)) {
|
||||
cat("\n[UNCERTAINLY LEVEL 2] (3) look for genus only, part of name\n")
|
||||
}
|
||||
if (nchar(g.x_backup_without_spp) > 4 & !b.x_trimmed %like% " ") {
|
||||
if (!grepl("^[A-Z][a-z]+", b.x_trimmed, ignore.case = FALSE)) {
|
||||
# not when input is like Genustext, because then Neospora would lead to Actinokineospora
|
||||
@ -1074,9 +1121,12 @@ exec_as.mo <- function(x,
|
||||
}
|
||||
|
||||
# (4) strip values between brackets ----
|
||||
if (isTRUE(debug)) {
|
||||
cat("\n[UNCERTAINLY LEVEL 2] (4) strip values between brackets\n")
|
||||
}
|
||||
a.x_backup_stripped <- gsub("( *[(].*[)] *)", " ", a.x_backup)
|
||||
a.x_backup_stripped <- trimws(gsub(" +", " ", a.x_backup_stripped))
|
||||
found <- suppressMessages(suppressWarnings(exec_as.mo(a.x_backup_stripped, initial_search = FALSE, allow_uncertain = FALSE)))
|
||||
found <- suppressMessages(suppressWarnings(exec_as.mo(a.x_backup_stripped, initial_search = FALSE, allow_uncertain = FALSE, debug = debug)))
|
||||
if (!empty_result(found) & nchar(g.x_backup_without_spp) >= 6) {
|
||||
found_result <- found
|
||||
found <- microorganismsDT[mo == found, ..property][[1]]
|
||||
@ -1092,6 +1142,9 @@ exec_as.mo <- function(x,
|
||||
}
|
||||
|
||||
# (5a) try to strip off half an element from end and check the remains ----
|
||||
if (isTRUE(debug)) {
|
||||
cat("\n[UNCERTAINLY LEVEL 2] (5a) try to strip off half an element from end and check the remains\n")
|
||||
}
|
||||
x_strip <- a.x_backup %>% strsplit(" ") %>% unlist()
|
||||
if (length(x_strip) > 1) {
|
||||
for (i in 1:(length(x_strip) - 1)) {
|
||||
@ -1100,7 +1153,7 @@ exec_as.mo <- function(x,
|
||||
# remove last half of the second term
|
||||
x_strip_collapsed <- paste(c(x_strip[1:(length(x_strip) - i)], lastword_half), collapse = " ")
|
||||
if (nchar(x_strip_collapsed) >= 4) {
|
||||
found <- suppressMessages(suppressWarnings(exec_as.mo(x_strip_collapsed, initial_search = FALSE, allow_uncertain = FALSE)))
|
||||
found <- suppressMessages(suppressWarnings(exec_as.mo(x_strip_collapsed, initial_search = FALSE, allow_uncertain = FALSE, debug = debug)))
|
||||
if (!empty_result(found)) {
|
||||
found_result <- found
|
||||
found <- microorganismsDT[mo == found, ..property][[1]]
|
||||
@ -1118,11 +1171,14 @@ exec_as.mo <- function(x,
|
||||
}
|
||||
}
|
||||
# (5b) try to strip off one element from end and check the remains ----
|
||||
if (isTRUE(debug)) {
|
||||
cat("\n[UNCERTAINLY LEVEL 2] (5b) try to strip off one element from end and check the remains\n")
|
||||
}
|
||||
if (length(x_strip) > 1) {
|
||||
for (i in 1:(length(x_strip) - 1)) {
|
||||
x_strip_collapsed <- paste(x_strip[1:(length(x_strip) - i)], collapse = " ")
|
||||
if (nchar(x_strip_collapsed) >= 4) {
|
||||
found <- suppressMessages(suppressWarnings(exec_as.mo(x_strip_collapsed, initial_search = FALSE, allow_uncertain = FALSE)))
|
||||
found <- suppressMessages(suppressWarnings(exec_as.mo(x_strip_collapsed, initial_search = FALSE, allow_uncertain = FALSE, debug = debug)))
|
||||
if (!empty_result(found)) {
|
||||
found_result <- found
|
||||
found <- microorganismsDT[mo == found, ..property][[1]]
|
||||
@ -1139,12 +1195,47 @@ exec_as.mo <- function(x,
|
||||
}
|
||||
}
|
||||
}
|
||||
# (5c) check for unknown yeasts/fungi ----
|
||||
if (isTRUE(debug)) {
|
||||
cat("\n[UNCERTAINLY LEVEL 2] (5b) check for unknown yeasts/fungi\n")
|
||||
}
|
||||
if (b.x_trimmed %like% "yeast") {
|
||||
found <- "F_YEAST"
|
||||
found_result <- found
|
||||
found <- microorganismsDT[mo == found, ..property][[1]]
|
||||
uncertainties <<- rbind(uncertainties,
|
||||
data.frame(uncertainty = 2,
|
||||
input = a.x_backup,
|
||||
fullname = microorganismsDT[mo == found_result[1L], fullname][[1]],
|
||||
mo = found_result[1L]))
|
||||
if (initial_search == TRUE) {
|
||||
set_mo_history(a.x_backup, get_mo_code(found[1L], property), 2, force = force_mo_history)
|
||||
}
|
||||
return(found[1L])
|
||||
}
|
||||
if (b.x_trimmed %like% "fungus") {
|
||||
found <- "F_FUNGUS"
|
||||
found_result <- found
|
||||
found <- microorganismsDT[mo == found, ..property][[1]]
|
||||
uncertainties <<- rbind(uncertainties,
|
||||
data.frame(uncertainty = 2,
|
||||
input = a.x_backup,
|
||||
fullname = microorganismsDT[mo == found_result[1L], fullname][[1]],
|
||||
mo = found_result[1L]))
|
||||
if (initial_search == TRUE) {
|
||||
set_mo_history(a.x_backup, get_mo_code(found[1L], property), 2, force = force_mo_history)
|
||||
}
|
||||
return(found[1L])
|
||||
}
|
||||
# (6) try to strip off one element from start and check the remains (only allow >= 2-part name outcome) ----
|
||||
if (isTRUE(debug)) {
|
||||
cat("\n[UNCERTAINLY LEVEL 2] (6) try to strip off one element from start and check the remains (only allow >= 2-part name outcome)\n")
|
||||
}
|
||||
x_strip <- a.x_backup %>% strsplit(" ") %>% unlist()
|
||||
if (length(x_strip) > 1 & nchar(g.x_backup_without_spp) >= 6) {
|
||||
for (i in 2:(length(x_strip))) {
|
||||
x_strip_collapsed <- paste(x_strip[i:length(x_strip)], collapse = " ")
|
||||
found <- suppressMessages(suppressWarnings(exec_as.mo(x_strip_collapsed, initial_search = FALSE, allow_uncertain = FALSE)))
|
||||
found <- suppressMessages(suppressWarnings(exec_as.mo(x_strip_collapsed, initial_search = FALSE, allow_uncertain = FALSE, debug = debug)))
|
||||
if (!empty_result(found)) {
|
||||
found_result <- found
|
||||
found <- microorganismsDT[mo == found_result[1L], ..property][[1]]
|
||||
@ -1167,11 +1258,14 @@ exec_as.mo <- function(x,
|
||||
|
||||
if (uncertainty_level >= 3) {
|
||||
# (7) try to strip off one element from start and check the remains ----
|
||||
if (isTRUE(debug)) {
|
||||
cat("\n[UNCERTAINLY LEVEL 3] (7) try to strip off one element from start and check the remains\n")
|
||||
}
|
||||
x_strip <- a.x_backup %>% strsplit(" ") %>% unlist()
|
||||
if (length(x_strip) > 1 & nchar(g.x_backup_without_spp) >= 6) {
|
||||
for (i in 2:(length(x_strip))) {
|
||||
x_strip_collapsed <- paste(x_strip[i:length(x_strip)], collapse = " ")
|
||||
found <- suppressMessages(suppressWarnings(exec_as.mo(x_strip_collapsed, initial_search = FALSE, allow_uncertain = FALSE)))
|
||||
found <- suppressMessages(suppressWarnings(exec_as.mo(x_strip_collapsed, initial_search = FALSE, allow_uncertain = FALSE, debug = debug)))
|
||||
if (!empty_result(found)) {
|
||||
found_result <- found
|
||||
found <- microorganismsDT[mo == found, ..property][[1]]
|
||||
@ -1189,6 +1283,9 @@ exec_as.mo <- function(x,
|
||||
}
|
||||
|
||||
# (8) part of a name (very unlikely match) ----
|
||||
if (isTRUE(debug)) {
|
||||
cat("\n[UNCERTAINLY LEVEL 3] (8) part of a name (very unlikely match)\n")
|
||||
}
|
||||
found <- microorganismsDT[fullname %like% f.x_withspaces_end_only]
|
||||
if (nrow(found) > 0) {
|
||||
found_result <- found[["mo"]]
|
||||
@ -1217,7 +1314,7 @@ exec_as.mo <- function(x,
|
||||
x_withspaces_end_only[i],
|
||||
x_backup_without_spp[i])
|
||||
if (!empty_result(x[i])) {
|
||||
# no set_mo_history: is already set in uncertain_fn()
|
||||
# no set_mo_history here - it is already set in uncertain_fn()
|
||||
next
|
||||
}
|
||||
|
||||
@ -1238,10 +1335,10 @@ exec_as.mo <- function(x,
|
||||
if (n_distinct(failures) > 1) {
|
||||
plural <- c("values", "them", "were")
|
||||
}
|
||||
total_failures <- length(x_input[x_input %in% failures & !x_input %in% c(NA, NULL, NaN)])
|
||||
total_failures <- length(x_input[as.character(x_input) %in% as.character(failures) & !x_input %in% c(NA, NULL, NaN)])
|
||||
total_n <- length(x_input[!x_input %in% c(NA, NULL, NaN)])
|
||||
msg <- paste0(nr2char(n_distinct(failures)), " unique ", plural[1],
|
||||
" (^= ", percent(total_failures / total_n, round = 1, force_zero = TRUE),
|
||||
" (covering ", percent(total_failures / total_n, round = 1, force_zero = TRUE),
|
||||
") could not be coerced and ", plural[3], " considered 'unknown'")
|
||||
if (n_distinct(failures) <= 10) {
|
||||
msg <- paste0(msg, ": ", paste('"', unique(failures), '"', sep = "", collapse = ', '))
|
||||
@ -1412,6 +1509,7 @@ print.mo <- function(x, ...) {
|
||||
|
||||
#' @exportMethod summary.mo
|
||||
#' @importFrom dplyr n_distinct
|
||||
#' @importFrom clean freq top_freq
|
||||
#' @export
|
||||
#' @noRd
|
||||
summary.mo <- function(object, ...) {
|
||||
|
||||
@ -151,13 +151,18 @@ mo_shortname <- function(x, language = get_locale(), ...) {
|
||||
x.mo <- AMR::as.mo(x, ...)
|
||||
metadata <- get_mo_failures_uncertainties_renamed()
|
||||
|
||||
replace_empty <- function(x) {
|
||||
x[x == ""] <- "spp."
|
||||
x
|
||||
}
|
||||
|
||||
# get first char of genus and complete species in English
|
||||
shortnames <- paste0(substr(mo_genus(x.mo, language = NULL), 1, 1), ". ", mo_species(x.mo, language = NULL))
|
||||
|
||||
shortnames <- paste0(substr(mo_genus(x.mo, language = NULL), 1, 1), ". ", replace_empty(mo_species(x.mo, language = NULL)))
|
||||
|
||||
# exceptions for Staphylococci
|
||||
shortnames[shortnames == "S. coagulase-negative" ] <- "CoNS"
|
||||
shortnames[shortnames == "S. coagulase-positive" ] <- "CoPS"
|
||||
# exceptions for Streptococci
|
||||
# exceptions for Streptococci: Streptococcus Group A -> GAS
|
||||
shortnames[shortnames %like% "S. group [ABCDFGHK]"] <- paste0("G", gsub("S. group ([ABCDFGHK])", "\\1", shortnames[shortnames %like% "S. group [ABCDFGHK]"]), "S")
|
||||
|
||||
load_mo_failures_uncertainties_renamed(metadata)
|
||||
|
||||
@ -28,8 +28,8 @@
|
||||
#' @param year_max highest year to use in the prediction model, defaults to 10 years after today
|
||||
#' @param year_every unit of sequence between lowest year found in the data and \code{year_max}
|
||||
#' @param minimum minimal amount of available isolates per year to include. Years containing less observations will be estimated by the model.
|
||||
#' @param model the statistical model of choice. Defaults to a generalised linear regression model with binomial distribution, assuming that a period of zero resistance was followed by a period of increasing resistance leading slowly to more and more resistance. See Details for valid options.
|
||||
#' @param I_as_S a logical to indicate whether values \code{I} should be treated as \code{S}
|
||||
#' @param model the statistical model of choice. Defaults to a generalised linear regression model with binomial distribution (i.e. using \code{\link{glm}(..., family = \link{binomial})}), assuming that a period of zero resistance was followed by a period of increasing resistance leading slowly to more and more resistance. See Details for valid options.
|
||||
#' @param I_as_S a logical to indicate whether values \code{I} should be treated as \code{S} (will otherwise be treated as \code{R})
|
||||
#' @param preserve_measurements a logical to indicate whether predictions of years that are actually available in the data should be overwritten by the original data. The standard errors of those years will be \code{NA}.
|
||||
#' @param info a logical to indicate whether textual analysis should be printed with the name and \code{\link{summary}} of the statistical model.
|
||||
#' @param main title of the plot
|
||||
@ -58,7 +58,7 @@
|
||||
#' @rdname resistance_predict
|
||||
#' @export
|
||||
#' @importFrom stats predict glm lm
|
||||
#' @importFrom dplyr %>% pull mutate mutate_at n group_by_at summarise filter filter_at all_vars n_distinct arrange case_when n_groups transmute
|
||||
#' @importFrom dplyr %>% pull mutate mutate_at n group_by_at summarise filter filter_at all_vars n_distinct arrange case_when n_groups transmute ungroup
|
||||
#' @inheritSection AMR Read more on our website!
|
||||
#' @examples
|
||||
#' x <- resistance_predict(septic_patients, col_ab = "AMX", year_min = 2010)
|
||||
@ -162,18 +162,19 @@ resistance_predict <- function(x,
|
||||
as.integer(format(as.Date(x), '%Y'))
|
||||
}
|
||||
}
|
||||
|
||||
|
||||
df <- x %>%
|
||||
mutate_at(col_ab, as.rsi) %>%
|
||||
mutate_at(col_ab, droplevels) %>%
|
||||
mutate_at(col_ab, ~(
|
||||
if (I_as_S == TRUE) {
|
||||
gsub("I", "S", .)
|
||||
} else {
|
||||
# then I as R
|
||||
gsub("I", "R", .)
|
||||
}
|
||||
)) %>%
|
||||
mutate_at(col_ab, droplevels)
|
||||
if (I_as_S == TRUE) {
|
||||
df <- df %>%
|
||||
mutate_at(col_ab, ~gsub("I", "S", .))
|
||||
} else {
|
||||
# then I as R
|
||||
df <- df %>%
|
||||
mutate_at(col_ab, ~gsub("I", "R", .))
|
||||
}
|
||||
df <- df %>%
|
||||
filter_at(col_ab, all_vars(!is.na(.))) %>%
|
||||
mutate(year = pull(., col_date) %>% year()) %>%
|
||||
group_by_at(c('year', col_ab)) %>%
|
||||
|
||||
4
R/rsi.R
@ -50,7 +50,7 @@
|
||||
#' @return Ordered factor with new class \code{rsi}
|
||||
#' @keywords rsi
|
||||
#' @export
|
||||
#' @importFrom dplyr %>%
|
||||
#' @importFrom dplyr %>% desc arrange filter
|
||||
#' @seealso \code{\link{as.mic}}
|
||||
#' @inheritSection AMR Read more on our website!
|
||||
#' @examples
|
||||
@ -73,6 +73,8 @@
|
||||
#'
|
||||
#' plot(rsi_data) # for percentages
|
||||
#' barplot(rsi_data) # for frequencies
|
||||
#'
|
||||
#' library(clean)
|
||||
#' freq(rsi_data) # frequency table with informative header
|
||||
#'
|
||||
#' # using dplyr's mutate
|
||||
|
||||
BIN
R/sysdata.rda
@ -24,7 +24,7 @@
|
||||
#' All antimicrobial drugs and their official names, ATC codes, ATC groups and defined daily dose (DDD) are included in this package, using the WHO Collaborating Centre for Drug Statistics Methodology.
|
||||
#' @section WHOCC:
|
||||
#' \if{html}{\figure{logo_who.png}{options: height=60px style=margin-bottom:5px} \cr}
|
||||
#' This package contains \strong{all ~450 antimicrobial drugs} and their Anatomical Therapeutic Chemical (ATC) codes, ATC groups and Defined Daily Dose (DDD) from the World Health Organization Collaborating Centre for Drug Statistics Methodology (WHOCC, \url{https://www.whocc.no}) and the Pharmaceuticals Community Register of the European Commission (\url{http://ec.europa.eu/health/documents/community-register/html/atc.htm}).
|
||||
#' This package contains \strong{all ~450 antimicrobial drugs} and their Anatomical Therapeutic Chemical (ATC) codes, ATC groups and Defined Daily Dose (DDD) from the World Health Organization Collaborating Centre for Drug Statistics Methodology (WHOCC, \url{https://www.whocc.no}) and the Pharmaceuticals Community Register of the European Commission (\url{http://ec.europa.eu/health/documents/community-register/html/atc.htm}). \strong{NOTE: The WHOCC copyright does not allow use for commercial purposes, unlike any other info from this package. See \url{https://www.whocc.no/copyright_disclaimer/}.}
|
||||
#'
|
||||
#' These have become the gold standard for international drug utilisation monitoring and research.
|
||||
#'
|
||||
|
||||
@ -62,12 +62,6 @@ navbar:
|
||||
- text: "Get properties of an antibiotic"
|
||||
icon: "fa-capsules"
|
||||
href: "reference/ab_property.html" # reference instead of article
|
||||
- text: "Create frequency tables"
|
||||
icon: "fa-sort-amount-down"
|
||||
href: "articles/freq.html"
|
||||
# - text: "Use the G-test"
|
||||
# icon: "fa-clipboard-check"
|
||||
# href: "reference/g.test.html" # reference instead of article
|
||||
- text: "Other: benchmarks"
|
||||
icon: "fa-shipping-fast"
|
||||
href: "articles/benchmarks.html"
|
||||
@ -130,13 +124,12 @@ reference:
|
||||
- title: "Analysing your data"
|
||||
desc: >
|
||||
Functions for conducting AMR analysis, like counting isolates, calculating
|
||||
resistance or susceptibility, creating frequency tables or make plots.
|
||||
resistance or susceptibility, or make plots.
|
||||
contents:
|
||||
- "`availability`"
|
||||
- "`count`"
|
||||
- "`portion`"
|
||||
- "`filter_ab_class`"
|
||||
- "`freq`"
|
||||
- "`g.test`"
|
||||
- "`ggplot_rsi`"
|
||||
- "`kurtosis`"
|
||||
|
||||
@ -1,4 +1,14 @@
|
||||
if_mo_property like_is_one_of this_value and_these_antibiotics have_these_values then_change_these_antibiotics to_value reference.rule reference.rule_group
|
||||
# ---------------------------------------------------------------------------------------------------
|
||||
# For editing this EUCAST reference file, these values can all be used for target antibiotics:
|
||||
# all_betalactams, aminoglycosides, carbapenems, cephalosporins, cephalosporins_without_CAZ, fluoroquinolones,
|
||||
# glycopeptides, macrolides, minopenicillins, polymyxins, streptogramins, tetracyclines, ureidopenicillins
|
||||
# and all separate EARS-Net letter codes like AMC. They can be separated by comma: 'AMC, fluoroquinolones'.
|
||||
# The if_mo_property column can be any column name from the AMR::microorganisms data set, or "genus_species" or "gramstain".
|
||||
# The like.is.one_of column must contain one of: like, is, one_of ('like' will read the first column as regular expression)
|
||||
# The EUCAST guideline contains references to the 'Burkholderia cepacia complex'. All species in this group can be found in: LiPuma J, Curr Opin Pulm Med. 2005 Nov;11(6):528-33. (PMID 16217180).
|
||||
# >>>>> IF YOU WANT TO IMPORT THIS FILE INTO YOUR OWN SOFTWARE, HAVE THE FIRST 10 LINES SKIPPED <<<<<
|
||||
# ---------------------------------------------------------------------------------------------------
|
||||
if_mo_property like.is.one_of this_value and_these_antibiotics have_these_values then_change_these_antibiotics to_value reference.rule reference.rule_group
|
||||
order is Enterobacteriales AMP S AMX S Enterobacteriales (Order) Breakpoints
|
||||
order is Enterobacteriales AMP I AMX I Enterobacteriales (Order) Breakpoints
|
||||
order is Enterobacteriales AMP R AMX R Enterobacteriales (Order) Breakpoints
|
||||
|
||||
|
Can't render this file because it contains an unexpected character in line 6 and column 94.
|
@ -1,14 +1,16 @@
|
||||
# EUCAST rules ----
|
||||
# For editing the reference file, these values can all be used for target antibiotics:
|
||||
# "aminoglycosides", "tetracyclines", "polymyxins", "macrolides", "glycopeptides",
|
||||
# "streptogramins", "cephalosporins", "cephalosporins_without_CAZ", "carbapenems",
|
||||
# "minopenicillins", "ureidopenicillins", "fluoroquinolones", "all_betalactams",
|
||||
# and all separate EARS-Net letter codes like "AMC". They can be separated by comma: "AMC, fluoroquinolones".
|
||||
# The mo_property can be any column name from the AMR::microorganisms data set, or "genus_species" or "gramstain".
|
||||
# The EUCAST guideline contains references to the 'Burkholderia cepacia complex'. The species in this group can be found in:
|
||||
# LiPuma JJ, Curr Opin Pulm Med. 2005 Nov;11(6):528-33. (PMID 16217180).
|
||||
# ---------------------------------------------------------------------------------------------------
|
||||
# For editing this EUCAST reference file, these values can all be used for target antibiotics:
|
||||
# all_betalactams, aminoglycosides, carbapenems, cephalosporins, cephalosporins_without_CAZ, fluoroquinolones,
|
||||
# glycopeptides, macrolides, minopenicillins, polymyxins, streptogramins, tetracyclines, ureidopenicillins
|
||||
# and all separate EARS-Net letter codes like AMC. They can be separated by comma: 'AMC, fluoroquinolones'.
|
||||
# The if_mo_property column can be any column name from the AMR::microorganisms data set, or "genus_species" or "gramstain".
|
||||
# The EUCAST guideline contains references to the 'Burkholderia cepacia complex'. All species in this group can be found in:
|
||||
# LiPuma J, Curr Opin Pulm Med. 2005 Nov;11(6):528-33. (PMID 16217180).
|
||||
# >>>>> IF YOU WANT TO IMPORT THIS FILE INTO YOUR OWN SOFTWARE, HAVE THE FIRST 10 LINES SKIPPED <<<<<
|
||||
# ---------------------------------------------------------------------------------------------------
|
||||
eucast_rules_file <- dplyr::arrange(
|
||||
.data = utils::read.delim(file = "data-raw/eucast_rules.tsv",
|
||||
skip = 10,
|
||||
sep = "\t",
|
||||
stringsAsFactors = FALSE,
|
||||
header = TRUE,
|
||||
|
||||
@ -99,7 +99,7 @@ MOs <- data_total %>%
|
||||
# and not all fungi: Aspergillus, Candida, Trichphyton and Pneumocystis are the most important,
|
||||
# so only keep these orders from the fungi:
|
||||
& !(kingdom == "Fungi"
|
||||
& !order %in% c("Eurotiales", "Saccharomycetales", "Schizosaccharomycetales", "Tremellales", "Onygenales", "Pneumocystales"))
|
||||
& !order %in% c("Eurotiales", "Mucorales", "Saccharomycetales", "Schizosaccharomycetales", "Tremellales", "Onygenales", "Pneumocystales"))
|
||||
)
|
||||
# or the genus has to be one of the genera we found in our hospitals last decades (Northern Netherlands, 2002-2018)
|
||||
| genus %in% c("Absidia", "Acremonium", "Actinotignum", "Alternaria", "Anaerosalibacter", "Ancylostoma", "Anisakis", "Apophysomyces",
|
||||
@ -123,7 +123,7 @@ MOs <- MOs %>%
|
||||
|
||||
MOs <- MOs %>%
|
||||
# remove text if it contains 'Not assigned' like phylum in viruses
|
||||
mutate_all(~gsub("Not assigned", "", .))
|
||||
mutate_all(~gsub("(Not assigned|\\[homonym\\]|\\[mistake\\])", "", ., ignore.case = TRUE))
|
||||
|
||||
MOs <- MOs %>%
|
||||
# Only keep first author, e.g. transform 'Smith, Jones, 2011' to 'Smith et al., 2011':
|
||||
@ -166,8 +166,10 @@ MOs <- MOs %>%
|
||||
|
||||
# Remove non-ASCII characters (these are not allowed by CRAN)
|
||||
MOs <- MOs %>%
|
||||
lapply(iconv, from = "UTF-8", to = "ASCII//TRANSLIT") %>%
|
||||
as_tibble(stringsAsFactors = FALSE)
|
||||
lapply(iconv, from = "UTF-8", to = "ASCII//TRANSLIT") %>%
|
||||
as_tibble(stringsAsFactors = FALSE) %>%
|
||||
# remove invalid characters
|
||||
mutate_all(~gsub("[\"'`]+", "", .))
|
||||
|
||||
# Split old taxonomic names - they refer in the original data to a new `taxonID` with `acceptedNameUsageID`
|
||||
MOs.old <- MOs %>%
|
||||
@ -219,6 +221,9 @@ MOs <- MOs %>%
|
||||
!(source == "DSMZ" & fullname %in% (MOs %>% filter(source == "CoL") %>% pull(fullname)))) %>%
|
||||
distinct(fullname, .keep_all = TRUE)
|
||||
|
||||
# what characters are in the fullnames?
|
||||
paste(unique(sort(unlist(strsplit(x = paste(MOs$fullname, collapse = ""), split = "")))), collapse = "")
|
||||
|
||||
# Add abbreviations so we can easily know which ones are which ones.
|
||||
# These will become valid and unique microbial IDs for the AMR package.
|
||||
MOs <- MOs %>%
|
||||
@ -295,7 +300,6 @@ MOs <- MOs %>%
|
||||
# put `mo` in front, followed by the rest
|
||||
select(mo, everything(), -abbr_other, -abbr_genus, -abbr_species, -abbr_subspecies)
|
||||
|
||||
|
||||
# add non-taxonomic entries
|
||||
MOs <- MOs %>%
|
||||
bind_rows(
|
||||
@ -348,6 +352,38 @@ MOs <- MOs %>%
|
||||
species_id = "",
|
||||
source = "manually added",
|
||||
stringsAsFactors = FALSE),
|
||||
data.frame(mo = "F_YEAST",
|
||||
col_id = NA_integer_,
|
||||
fullname = "(unknown yeast)",
|
||||
kingdom = "Fungi",
|
||||
phylum = "(unknown phylum)",
|
||||
class = "(unknown class)",
|
||||
order = "(unknown order)",
|
||||
family = "(unknown family)",
|
||||
genus = "(unknown genus)",
|
||||
species = "(unknown species)",
|
||||
subspecies = "(unknown subspecies)",
|
||||
rank = "species",
|
||||
ref = NA_character_,
|
||||
species_id = "",
|
||||
source = "manually added",
|
||||
stringsAsFactors = FALSE),
|
||||
data.frame(mo = "F_FUNGUS",
|
||||
col_id = NA_integer_,
|
||||
fullname = "(unknown fungus)",
|
||||
kingdom = "Fungi",
|
||||
phylum = "(unknown phylum)",
|
||||
class = "(unknown class)",
|
||||
order = "(unknown order)",
|
||||
family = "(unknown family)",
|
||||
genus = "(unknown genus)",
|
||||
species = "(unknown species)",
|
||||
subspecies = "(unknown subspecies)",
|
||||
rank = "species",
|
||||
ref = NA_character_,
|
||||
species_id = "",
|
||||
source = "manually added",
|
||||
stringsAsFactors = FALSE),
|
||||
# CoNS
|
||||
MOs %>%
|
||||
filter(genus == "Staphylococcus", species == "epidermidis") %>% .[1,] %>%
|
||||
@ -488,6 +524,11 @@ MOs <- MOs %>%
|
||||
sum(duplicated(MOs$mo))
|
||||
colnames(MOs)
|
||||
|
||||
# here we welcome the new ones:
|
||||
MOs %>% filter(!fullname %in% AMR::microorganisms$fullname) %>% View()
|
||||
# and the ones we lost:
|
||||
AMR::microorganisms %>% filter(!fullname %in% MOs$fullname) %>% View()
|
||||
|
||||
# set prevalence per species
|
||||
MOs <- MOs %>%
|
||||
mutate(prevalence = case_when(
|
||||
@ -534,12 +575,16 @@ MOs.old$col_id <- as.integer(MOs.old$col_id)
|
||||
MOs.old$col_id_new <- as.integer(MOs.old$col_id_new)
|
||||
|
||||
# save
|
||||
### for other server
|
||||
saveRDS(MOs, "microorganisms.rds")
|
||||
saveRDS(MOs.old, "microorganisms.old.rds")
|
||||
### for same server
|
||||
microorganisms <- MOs
|
||||
microorganisms.old <- MOs.old
|
||||
|
||||
# on the server, do:
|
||||
usethis::use_data(microorganisms, overwrite = TRUE, version = 2)
|
||||
usethis::use_data(microorganisms.old, overwrite = TRUE, version = 2)
|
||||
rm(microorganisms)
|
||||
rm(microorganisms.old)
|
||||
# and update the year in R/data.R
|
||||
# and update the year and dimensions in R/data.R
|
||||
|
||||
657
data-raw/reproduction_of_microorganisms_new.R
Normal file
@ -0,0 +1,657 @@
|
||||
# ---------------------------------------------------------------------------------
|
||||
# Reproduction of the `microorganisms` data set
|
||||
# ---------------------------------------------------------------------------------
|
||||
# Data retrieved from:
|
||||
#
|
||||
# [1] Catalogue of Life (CoL) through the Encyclopaedia of Life
|
||||
# https://opendata.eol.org/dataset/catalogue-of-life/
|
||||
# * Download the resource file with a name like "Catalogue of Life yyyy-mm-dd"
|
||||
# * Extract "taxon.tab"
|
||||
#
|
||||
# [2] Global Biodiversity Information Facility (GBIF)
|
||||
# https://doi.org/10.15468/39omei
|
||||
# * Extract "Taxon.tsv"
|
||||
#
|
||||
# [3] Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ)
|
||||
# https://www.dsmz.de/support/bacterial-nomenclature-up-to-date-downloads.html
|
||||
# * Download the latest "Complete List" as xlsx file (DSMZ_bactnames.xlsx)
|
||||
# ---------------------------------------------------------------------------------
|
||||
|
||||
library(dplyr)
|
||||
library(AMR)
|
||||
|
||||
data_col <- data.table::fread("Documents/taxon.tab")
|
||||
data_gbif <- data.table::fread("Documents/Taxon.tsv")
|
||||
|
||||
# read the xlsx file from DSMZ (only around 2.5 MB):
|
||||
data_dsmz <- readxl::read_xlsx("Downloads/DSMZ_bactnames.xlsx")
|
||||
|
||||
# the CoL data is over 3.7M rows:
|
||||
data_col %>% freq(kingdom)
|
||||
# Item Count Percent Cum. Count Cum. Percent
|
||||
# --- ---------- ---------- -------- ----------- -------------
|
||||
# 1 Animalia 2,225,627 59.1% 2,225,627 59.1%
|
||||
# 2 Plantae 1,177,412 31.3% 3,403,039 90.4%
|
||||
# 3 Fungi 290,145 7.7% 3,693,184 98.1%
|
||||
# 4 Chromista 47,126 1.3% 3,740,310 99.3%
|
||||
# 5 Bacteria 14,478 0.4% 3,754,788 99.7%
|
||||
# 6 Protozoa 6,060 0.2% 3,760,848 99.9%
|
||||
# 7 Viruses 3,827 0.1% 3,764,675 100.0%
|
||||
# 8 Archaea 610 0.0% 3,765,285 100.0%
|
||||
|
||||
# the GBIF data is over 5.8M rows:
|
||||
data_gbif %>% freq(kingdom)
|
||||
# Item Count Percent Cum. Count Cum. Percent
|
||||
# --- --------------- ---------- -------- ----------- -------------
|
||||
# 1 Animalia 3,264,138 55.7% 3,264,138 55.7%
|
||||
# 2 Plantae 1,814,962 31.0% 5,079,100 86.7%
|
||||
# 3 Fungi 538,086 9.2% 5,617,186 95.9%
|
||||
# 4 Chromista 181,374 3.1% 5,798,560 99.0%
|
||||
# 5 Bacteria 24,048 0.4% 5,822,608 99.4%
|
||||
# 6 Protozoa 15,138 0.3% 5,837,746 99.7%
|
||||
# 7 incertae sedis 9,995 0.2% 5,847,741 99.8%
|
||||
# 8 Viruses 9,630 0.2% 5,857,371 100.0%
|
||||
# 9 Archaea 771 0.0% 5,858,142 100.0%
|
||||
|
||||
|
||||
# Clean up helper function ------------------------------------------------
|
||||
clean_new <- function(new) {
|
||||
new %>%
|
||||
# only the ones that have no new ID to refer to a newer name
|
||||
filter(is.na(col_id_new)) %>%
|
||||
filter(
|
||||
(
|
||||
# we only want all MICROorganisms and no viruses
|
||||
!kingdom %in% c("Animalia", "Chromista", "Plantae", "Viruses")
|
||||
# and not all fungi: Aspergillus, Candida, Trichphyton and Pneumocystis are the most important,
|
||||
# so only keep these orders from the fungi:
|
||||
& !(kingdom == "Fungi"
|
||||
& !order %in% c("Eurotiales", "Saccharomycetales", "Schizosaccharomycetales", "Tremellales", "Onygenales", "Pneumocystales"))
|
||||
)
|
||||
# or the family has to contain a genus we found in our hospitals last decades (Northern Netherlands, 2002-2018)
|
||||
| genus %in% c("Absidia", "Acremonium", "Actinotignum", "Alternaria", "Anaerosalibacter", "Ancylostoma", "Anisakis", "Apophysomyces",
|
||||
"Arachnia", "Ascaris", "Aureobacterium", "Aureobasidium", "Balantidum", "Bilophilia", "Branhamella", "Brochontrix",
|
||||
"Brugia", "Calymmatobacterium", "Catabacter", "Chilomastix", "Chryseomonas", "Cladophialophora", "Cladosporium",
|
||||
"Clonorchis", "Cordylobia", "Curvularia", "Demodex", "Dermatobia", "Diphyllobothrium", "Dracunculus", "Echinococcus",
|
||||
"Enterobius", "Euascomycetes", "Exophiala", "Fasciola", "Fusarium", "Hendersonula", "Hymenolepis", "Kloeckera",
|
||||
"Koserella", "Larva", "Leishmania", "Lelliottia", "Loa", "Lumbricus", "Malassezia", "Metagonimus", "Molonomonas",
|
||||
"Mucor", "Nattrassia", "Necator", "Novospingobium", "Onchocerca", "Opistorchis", "Paragonimus", "Paramyxovirus",
|
||||
"Pediculus", "Phoma", "Phthirus", "Pityrosporum", "Pseudallescheria", "Pulex", "Rhizomucor", "Rhizopus", "Rhodotorula",
|
||||
"Salinococcus", "Sanguibacteroides", "Schistosoma", "Scopulariopsis", "Scytalidium", "Sporobolomyces", "Stomatococcus",
|
||||
"Strongyloides", "Syncephalastraceae", "Taenia", "Torulopsis", "Trichinella", "Trichobilharzia", "Trichomonas",
|
||||
"Trichosporon", "Trichuris", "Trypanosoma", "Wuchereria")) %>%
|
||||
mutate(
|
||||
authors2 = iconv(ref, from = "UTF-8", to = "ASCII//TRANSLIT"),
|
||||
# remove leading and trailing brackets
|
||||
authors2 = gsub("^[(](.*)[)]$", "\\1", authors2),
|
||||
# only take part after brackets if there's a name
|
||||
authors2 = ifelse(grepl(".*[)] [a-zA-Z]+.*", authors2),
|
||||
gsub(".*[)] (.*)", "\\1", authors2),
|
||||
authors2),
|
||||
# get year from last 4 digits
|
||||
lastyear = as.integer(gsub(".*([0-9]{4})$", "\\1", authors2)),
|
||||
# can never be later than now
|
||||
lastyear = ifelse(lastyear > as.integer(format(Sys.Date(), "%Y")),
|
||||
NA,
|
||||
lastyear),
|
||||
# get authors without last year
|
||||
authors = gsub("(.*)[0-9]{4}$", "\\1", authors2),
|
||||
# remove nonsense characters from names
|
||||
authors = gsub("[^a-zA-Z,'& -]", "", authors),
|
||||
# remove trailing and leading spaces
|
||||
authors = trimws(authors),
|
||||
# only keep first author and replace all others by 'et al'
|
||||
authors = gsub("(,| and| et| &| ex| emend\\.?) .*", " et al.", authors),
|
||||
# et al. always with ending dot
|
||||
authors = gsub(" et al\\.?", " et al.", authors),
|
||||
authors = gsub(" ?,$", "", authors),
|
||||
# don't start with 'sensu' or 'ehrenb'
|
||||
authors = gsub("^(sensu|Ehrenb.?) ", "", authors, ignore.case = TRUE),
|
||||
# no initials, only surname
|
||||
authors = gsub("^([A-Z]+ )+", "", authors, ignore.case = FALSE),
|
||||
# combine author and year if year is available
|
||||
ref = ifelse(!is.na(lastyear),
|
||||
paste0(authors, ", ", lastyear),
|
||||
authors),
|
||||
# fix beginning and ending
|
||||
ref = gsub(", $", "", ref),
|
||||
ref = gsub("^, ", "", ref)) %>%
|
||||
# remove text if it contains 'Not assigned' like phylum in viruses
|
||||
mutate_all(~gsub("Not assigned", "", .)) %>%
|
||||
# Remove non-ASCII characters (these are not allowed by CRAN)
|
||||
lapply(iconv, from = "UTF-8", to = "ASCII//TRANSLIT") %>%
|
||||
as_tibble(stringsAsFactors = FALSE) %>%
|
||||
mutate(fullname = trimws(case_when(rank == "family" ~ family,
|
||||
rank == "order" ~ order,
|
||||
rank == "class" ~ class,
|
||||
rank == "phylum" ~ phylum,
|
||||
rank == "kingdom" ~ kingdom,
|
||||
TRUE ~ paste(genus, species, subspecies))))
|
||||
}
|
||||
clean_old <- function(old, new) {
|
||||
old %>%
|
||||
# only the ones that exist in the new data set
|
||||
filter(col_id_new %in% new$col_id) %>%
|
||||
mutate(
|
||||
authors2 = iconv(ref, from = "UTF-8", to = "ASCII//TRANSLIT"),
|
||||
# remove leading and trailing brackets
|
||||
authors2 = gsub("^[(](.*)[)]$", "\\1", authors2),
|
||||
# only take part after brackets if there's a name
|
||||
authors2 = ifelse(grepl(".*[)] [a-zA-Z]+.*", authors2),
|
||||
gsub(".*[)] (.*)", "\\1", authors2),
|
||||
authors2),
|
||||
# get year from last 4 digits
|
||||
lastyear = as.integer(gsub(".*([0-9]{4})$", "\\1", authors2)),
|
||||
# can never be later than now
|
||||
lastyear = ifelse(lastyear > as.integer(format(Sys.Date(), "%Y")),
|
||||
NA,
|
||||
lastyear),
|
||||
# get authors without last year
|
||||
authors = gsub("(.*)[0-9]{4}$", "\\1", authors2),
|
||||
# remove nonsense characters from names
|
||||
authors = gsub("[^a-zA-Z,'& -]", "", authors),
|
||||
# remove trailing and leading spaces
|
||||
authors = trimws(authors),
|
||||
# only keep first author and replace all others by 'et al'
|
||||
authors = gsub("(,| and| et| &| ex| emend\\.?) .*", " et al.", authors),
|
||||
# et al. always with ending dot
|
||||
authors = gsub(" et al\\.?", " et al.", authors),
|
||||
authors = gsub(" ?,$", "", authors),
|
||||
# don't start with 'sensu' or 'ehrenb'
|
||||
authors = gsub("^(sensu|Ehrenb.?) ", "", authors, ignore.case = TRUE),
|
||||
# no initials, only surname
|
||||
authors = gsub("^([A-Z]+ )+", "", authors, ignore.case = FALSE),
|
||||
# combine author and year if year is available
|
||||
ref = ifelse(!is.na(lastyear),
|
||||
paste0(authors, ", ", lastyear),
|
||||
authors),
|
||||
# fix beginning and ending
|
||||
ref = gsub(", $", "", ref),
|
||||
ref = gsub("^, ", "", ref)) %>%
|
||||
# remove text if it contains 'Not assigned' like phylum in viruses
|
||||
mutate_all(~gsub("Not assigned", "", .)) %>%
|
||||
# Remove non-ASCII characters (these are not allowed by CRAN)
|
||||
lapply(iconv, from = "UTF-8", to = "ASCII//TRANSLIT") %>%
|
||||
as_tibble(stringsAsFactors = FALSE) %>%
|
||||
select(col_id_new, fullname, ref, authors2) %>%
|
||||
left_join(new %>% select(col_id, fullname_new = fullname), by = c(col_id_new = "col_id")) %>%
|
||||
mutate(fullname = trimws(
|
||||
gsub("(.*)[(].*", "\\1",
|
||||
stringr::str_replace(
|
||||
string = fullname,
|
||||
pattern = stringr::fixed(authors2),
|
||||
replacement = "")) %>%
|
||||
gsub(" (var|f|subsp)[.]", "", .))) %>%
|
||||
select(-c("col_id_new", "authors2")) %>%
|
||||
filter(!is.na(fullname), !is.na(fullname_new)) %>%
|
||||
filter(fullname != fullname_new, !fullname %like% "^[?]")
|
||||
}
|
||||
|
||||
# clean CoL and GBIF ----
|
||||
# clean data_col
|
||||
data_col <- data_col %>%
|
||||
as_tibble() %>%
|
||||
select(col_id = taxonID,
|
||||
col_id_new = acceptedNameUsageID,
|
||||
fullname = scientificName,
|
||||
kingdom,
|
||||
phylum,
|
||||
class,
|
||||
order,
|
||||
family,
|
||||
genus,
|
||||
species = specificEpithet,
|
||||
subspecies = infraspecificEpithet,
|
||||
rank = taxonRank,
|
||||
ref = scientificNameAuthorship,
|
||||
species_id = furtherInformationURL) %>%
|
||||
mutate(source = "CoL")
|
||||
# split into old and new
|
||||
data_col.new <- data_col %>% clean_new()
|
||||
data_col.old <- data_col %>% clean_old(new = data_col.new)
|
||||
rm(data_col)
|
||||
|
||||
# clean data_gbif
|
||||
data_gbif <- data_gbif %>%
|
||||
as_tibble() %>%
|
||||
filter(
|
||||
# no uncertain taxonomic placements
|
||||
taxonRemarks != "doubtful",
|
||||
kingdom != "incertae sedis",
|
||||
taxonRank != "unranked") %>%
|
||||
transmute(col_id = taxonID,
|
||||
col_id_new = acceptedNameUsageID,
|
||||
fullname = scientificName,
|
||||
kingdom,
|
||||
phylum,
|
||||
class,
|
||||
order,
|
||||
family,
|
||||
genus,
|
||||
species = specificEpithet,
|
||||
subspecies = infraspecificEpithet,
|
||||
rank = taxonRank,
|
||||
ref = scientificNameAuthorship,
|
||||
species_id = as.character(parentNameUsageID)) %>%
|
||||
mutate(source = "GBIF")
|
||||
# split into old and new
|
||||
data_gbif.new <- data_gbif %>% clean_new()
|
||||
data_gbif.old <- data_gbif %>% clean_old(new = data_gbif.new)
|
||||
rm(data_gbif)
|
||||
|
||||
# put CoL and GBIF together ----
|
||||
MOs.new <- bind_rows(data_col.new,
|
||||
data_gbif.new) %>%
|
||||
mutate(taxonomic_tree_length = nchar(trimws(paste(kingdom, phylum, class, order, family, genus, species, subspecies)))) %>%
|
||||
arrange(desc(taxonomic_tree_length)) %>%
|
||||
distinct(fullname, .keep_all = TRUE) %>%
|
||||
select(-c("col_id_new", "authors2", "authors", "lastyear", "taxonomic_tree_length")) %>%
|
||||
arrange(fullname)
|
||||
MOs.old <- bind_rows(data_col.old,
|
||||
data_gbif.old) %>%
|
||||
distinct(fullname, .keep_all = TRUE) %>%
|
||||
arrange(fullname)
|
||||
|
||||
# clean up DSMZ ---
|
||||
data_dsmz <- data_dsmz %>%
|
||||
as_tibble() %>%
|
||||
transmute(col_id = NA_integer_,
|
||||
col_id_new = NA_integer_,
|
||||
fullname = "",
|
||||
# kingdom = "",
|
||||
# phylum = "",
|
||||
# class = "",
|
||||
# order = "",
|
||||
# family = "",
|
||||
genus = ifelse(is.na(GENUS), "", GENUS),
|
||||
species = ifelse(is.na(SPECIES), "", SPECIES),
|
||||
subspecies = ifelse(is.na(SUBSPECIES), "", SUBSPECIES),
|
||||
rank = ifelse(species == "", "genus", "species"),
|
||||
ref = AUTHORS,
|
||||
species_id = as.character(RECORD_NO),
|
||||
source = "DSMZ")
|
||||
|
||||
# DSMZ only contains genus/(sub)species, try to find taxonomic properties based on genus and data_col
|
||||
ref_taxonomy <- MOs.new %>%
|
||||
distinct(genus, .keep_all = TRUE) %>%
|
||||
filter(family != "") %>%
|
||||
filter(genus %in% data_dsmz$genus) %>%
|
||||
distinct(genus, .keep_all = TRUE) %>%
|
||||
select(kingdom, phylum, class, order, family, genus)
|
||||
|
||||
data_dsmz <- data_dsmz %>%
|
||||
left_join(ref_taxonomy, by = "genus") %>%
|
||||
mutate(kingdom = "Bacteria")
|
||||
|
||||
data_dsmz.new <- data_dsmz %>%
|
||||
clean_new() %>%
|
||||
distinct(fullname, .keep_all = TRUE) %>%
|
||||
select(colnames(MOs.new)) %>%
|
||||
arrange(fullname)
|
||||
|
||||
# combine everything ----
|
||||
MOs <- bind_rows(MOs.new,
|
||||
data_dsmz.new) %>%
|
||||
distinct(fullname, .keep_all = TRUE) %>%
|
||||
# not the ones that are old
|
||||
filter(!fullname %in% MOs.old$fullname) %>%
|
||||
arrange(fullname) %>%
|
||||
mutate(col_id = ifelse(source != "CoL", NA_integer_, col_id)) %>%
|
||||
filter(fullname != "")
|
||||
|
||||
rm(data_col.new)
|
||||
rm(data_col.old)
|
||||
rm(data_gbif.new)
|
||||
rm(data_gbif.old)
|
||||
rm(data_dsmz)
|
||||
rm(data_dsmz.new)
|
||||
rm(ref_taxonomy)
|
||||
rm(MOs.new)
|
||||
|
||||
MOs.bak <- MOs
|
||||
|
||||
# Trichomonas trick ----
|
||||
# for species in Trypanosoma and Trichomonas we observe al lot of taxonomic info missing
|
||||
MOs %>% filter(genus %in% c("Trypanosoma", "Trichomonas")) %>% View()
|
||||
MOs[which(MOs$genus == "Trypanosoma"), "kingdom"] <- MOs[which(MOs$fullname == "Trypanosoma"),]$kingdom
|
||||
MOs[which(MOs$genus == "Trypanosoma"), "phylum"] <- MOs[which(MOs$fullname == "Trypanosoma"),]$phylum
|
||||
MOs[which(MOs$genus == "Trypanosoma"), "class"] <- MOs[which(MOs$fullname == "Trypanosoma"),]$class
|
||||
MOs[which(MOs$genus == "Trypanosoma"), "order"] <- MOs[which(MOs$fullname == "Trypanosoma"),]$order
|
||||
MOs[which(MOs$genus == "Trypanosoma"), "family"] <- MOs[which(MOs$fullname == "Trypanosoma"),]$family
|
||||
MOs[which(MOs$genus == "Trichomonas"), "kingdom"] <- MOs[which(MOs$fullname == "Trichomonas"),]$kingdom
|
||||
MOs[which(MOs$genus == "Trichomonas"), "phylum"] <- MOs[which(MOs$fullname == "Trichomonas"),]$phylum
|
||||
MOs[which(MOs$genus == "Trichomonas"), "class"] <- MOs[which(MOs$fullname == "Trichomonas"),]$class
|
||||
MOs[which(MOs$genus == "Trichomonas"), "order"] <- MOs[which(MOs$fullname == "Trichomonas"),]$order
|
||||
MOs[which(MOs$genus == "Trichomonas"), "family"] <- MOs[which(MOs$fullname == "Trichomonas"),]$family
|
||||
|
||||
# fill taxonomic properties that are missing
|
||||
MOs <- MOs %>%
|
||||
mutate(phylum = ifelse(phylum %in% c(NA, ""), "(unknown phylum)", phylum),
|
||||
class = ifelse(class %in% c(NA, ""), "(unknown class)", class),
|
||||
order = ifelse(order %in% c(NA, ""), "(unknown order)", order),
|
||||
family = ifelse(family %in% c(NA, ""), "(unknown family)", family))
|
||||
|
||||
# Abbreviations ----
|
||||
# Add abbreviations so we can easily know which ones are which ones.
|
||||
# These will become valid and unique microbial IDs for the AMR package.
|
||||
MOs <- MOs %>%
|
||||
arrange(kingdom, fullname) %>%
|
||||
group_by(kingdom) %>%
|
||||
mutate(abbr_other = case_when(
|
||||
rank == "family" ~ paste0("[FAM]_",
|
||||
abbreviate(family,
|
||||
minlength = 8,
|
||||
use.classes = TRUE,
|
||||
method = "both.sides",
|
||||
strict = FALSE)),
|
||||
rank == "order" ~ paste0("[ORD]_",
|
||||
abbreviate(order,
|
||||
minlength = 8,
|
||||
use.classes = TRUE,
|
||||
method = "both.sides",
|
||||
strict = FALSE)),
|
||||
rank == "class" ~ paste0("[CLS]_",
|
||||
abbreviate(class,
|
||||
minlength = 8,
|
||||
use.classes = TRUE,
|
||||
method = "both.sides",
|
||||
strict = FALSE)),
|
||||
rank == "phylum" ~ paste0("[PHL]_",
|
||||
abbreviate(phylum,
|
||||
minlength = 8,
|
||||
use.classes = TRUE,
|
||||
method = "both.sides",
|
||||
strict = FALSE)),
|
||||
rank == "kingdom" ~ paste0("[KNG]_", kingdom),
|
||||
TRUE ~ NA_character_
|
||||
)) %>%
|
||||
# abbreviations determined per kingdom and family
|
||||
# becuase they are part of the abbreviation
|
||||
mutate(abbr_genus = abbreviate(genus,
|
||||
minlength = 7,
|
||||
use.classes = TRUE,
|
||||
method = "both.sides",
|
||||
strict = FALSE)) %>%
|
||||
ungroup() %>%
|
||||
group_by(genus) %>%
|
||||
# species abbreviations may be the same between genera
|
||||
# because the genus abbreviation is part of the abbreviation
|
||||
mutate(abbr_species = abbreviate(stringr::str_to_title(species),
|
||||
minlength = 3,
|
||||
use.classes = FALSE,
|
||||
method = "both.sides")) %>%
|
||||
ungroup() %>%
|
||||
group_by(genus, species) %>%
|
||||
mutate(abbr_subspecies = abbreviate(stringr::str_to_title(subspecies),
|
||||
minlength = 3,
|
||||
use.classes = FALSE,
|
||||
method = "both.sides")) %>%
|
||||
ungroup() %>%
|
||||
# remove trailing underscores
|
||||
mutate(mo = gsub("_+$", "",
|
||||
toupper(paste(
|
||||
# first character: kingdom
|
||||
ifelse(kingdom %in% c("Animalia", "Plantae"),
|
||||
substr(kingdom, 1, 2),
|
||||
substr(kingdom, 1, 1)),
|
||||
# next: genus, species, subspecies
|
||||
ifelse(is.na(abbr_other),
|
||||
paste(abbr_genus,
|
||||
abbr_species,
|
||||
abbr_subspecies,
|
||||
sep = "_"),
|
||||
abbr_other),
|
||||
sep = "_")))) %>%
|
||||
mutate(mo = ifelse(duplicated(.$mo),
|
||||
# these one or two must be unique too
|
||||
paste0(mo, "1"),
|
||||
mo),
|
||||
fullname = ifelse(fullname == "",
|
||||
trimws(paste(genus, species, subspecies)),
|
||||
fullname)) %>%
|
||||
# put `mo` in front, followed by the rest
|
||||
select(mo, everything(), -abbr_other, -abbr_genus, -abbr_species, -abbr_subspecies)
|
||||
|
||||
# add non-taxonomic entries
|
||||
MOs <- MOs %>%
|
||||
bind_rows(
|
||||
# Unknowns
|
||||
data.frame(mo = "UNKNOWN",
|
||||
col_id = NA_integer_,
|
||||
fullname = "(unknown name)",
|
||||
kingdom = "(unknown kingdom)",
|
||||
phylum = "(unknown phylum)",
|
||||
class = "(unknown class)",
|
||||
order = "(unknown order)",
|
||||
family = "(unknown family)",
|
||||
genus = "(unknown genus)",
|
||||
species = "(unknown species)",
|
||||
subspecies = "(unknown subspecies)",
|
||||
rank = "(unknown rank)",
|
||||
ref = NA_character_,
|
||||
species_id = "",
|
||||
source = "manually added",
|
||||
stringsAsFactors = FALSE),
|
||||
data.frame(mo = "B_GRAMN",
|
||||
col_id = NA_integer_,
|
||||
fullname = "(unknown Gram-negatives)",
|
||||
kingdom = "Bacteria",
|
||||
phylum = "(unknown phylum)",
|
||||
class = "(unknown class)",
|
||||
order = "(unknown order)",
|
||||
family = "(unknown family)",
|
||||
genus = "(unknown Gram-negatives)",
|
||||
species = "(unknown species)",
|
||||
subspecies = "(unknown subspecies)",
|
||||
rank = "species",
|
||||
ref = NA_character_,
|
||||
species_id = "",
|
||||
source = "manually added",
|
||||
stringsAsFactors = FALSE),
|
||||
data.frame(mo = "B_GRAMP",
|
||||
col_id = NA_integer_,
|
||||
fullname = "(unknown Gram-positives)",
|
||||
kingdom = "Bacteria",
|
||||
phylum = "(unknown phylum)",
|
||||
class = "(unknown class)",
|
||||
order = "(unknown order)",
|
||||
family = "(unknown family)",
|
||||
genus = "(unknown Gram-positives)",
|
||||
species = "(unknown species)",
|
||||
subspecies = "(unknown subspecies)",
|
||||
rank = "species",
|
||||
ref = NA_character_,
|
||||
species_id = "",
|
||||
source = "manually added",
|
||||
stringsAsFactors = FALSE),
|
||||
# CoNS
|
||||
MOs %>%
|
||||
filter(genus == "Staphylococcus", species == "") %>% .[1,] %>%
|
||||
mutate(mo = paste(mo, "CNS", sep = "_"),
|
||||
rank = "species",
|
||||
col_id = NA_integer_,
|
||||
species = "coagulase-negative",
|
||||
fullname = "Coagulase-negative Staphylococcus (CoNS)",
|
||||
ref = NA_character_,
|
||||
species_id = "",
|
||||
source = "manually added"),
|
||||
# CoPS
|
||||
MOs %>%
|
||||
filter(genus == "Staphylococcus", species == "") %>% .[1,] %>%
|
||||
mutate(mo = paste(mo, "CPS", sep = "_"),
|
||||
rank = "species",
|
||||
col_id = NA_integer_,
|
||||
species = "coagulase-positive",
|
||||
fullname = "Coagulase-positive Staphylococcus (CoPS)",
|
||||
ref = NA_character_,
|
||||
species_id = "",
|
||||
source = "manually added"),
|
||||
# Streptococci groups A, B, C, F, H, K
|
||||
MOs %>%
|
||||
filter(genus == "Streptococcus", species == "pyogenes") %>% .[1,] %>%
|
||||
# we can keep all other details, since S. pyogenes is the only member of group A
|
||||
mutate(mo = paste(MOs[MOs$fullname == "Streptococcus",]$mo, "GRA", sep = "_"),
|
||||
species = "group A" ,
|
||||
fullname = "Streptococcus group A"),
|
||||
MOs %>%
|
||||
filter(genus == "Streptococcus", species == "agalactiae") %>% .[1,] %>%
|
||||
# we can keep all other details, since S. agalactiae is the only member of group B
|
||||
mutate(mo = paste(MOs[MOs$fullname == "Streptococcus",]$mo, "GRB", sep = "_"),
|
||||
species = "group B" ,
|
||||
fullname = "Streptococcus group B"),
|
||||
MOs %>%
|
||||
filter(genus == "Streptococcus", species == "dysgalactiae") %>% .[1,] %>%
|
||||
mutate(mo = paste(MOs[MOs$fullname == "Streptococcus",]$mo, "GRC", sep = "_"),
|
||||
col_id = NA_integer_,
|
||||
species = "group C" ,
|
||||
fullname = "Streptococcus group C",
|
||||
ref = NA_character_,
|
||||
species_id = "",
|
||||
source = "manually added"),
|
||||
MOs %>%
|
||||
filter(genus == "Streptococcus", species == "agalactiae") %>% .[1,] %>%
|
||||
mutate(mo = paste(MOs[MOs$fullname == "Streptococcus",]$mo, "GRD", sep = "_"),
|
||||
col_id = NA_integer_,
|
||||
species = "group D" ,
|
||||
fullname = "Streptococcus group D",
|
||||
ref = NA_character_,
|
||||
species_id = "",
|
||||
source = "manually added"),
|
||||
MOs %>%
|
||||
filter(genus == "Streptococcus", species == "agalactiae") %>% .[1,] %>%
|
||||
mutate(mo = paste(MOs[MOs$fullname == "Streptococcus",]$mo, "GRF", sep = "_"),
|
||||
col_id = NA_integer_,
|
||||
species = "group F" ,
|
||||
fullname = "Streptococcus group F",
|
||||
ref = NA_character_,
|
||||
species_id = "",
|
||||
source = "manually added"),
|
||||
MOs %>%
|
||||
filter(genus == "Streptococcus", species == "agalactiae") %>% .[1,] %>%
|
||||
mutate(mo = paste(MOs[MOs$fullname == "Streptococcus",]$mo, "GRG", sep = "_"),
|
||||
col_id = NA_integer_,
|
||||
species = "group G" ,
|
||||
fullname = "Streptococcus group G",
|
||||
ref = NA_character_,
|
||||
species_id = "",
|
||||
source = "manually added"),
|
||||
MOs %>%
|
||||
filter(genus == "Streptococcus", species == "agalactiae") %>% .[1,] %>%
|
||||
mutate(mo = paste(MOs[MOs$fullname == "Streptococcus",]$mo, "GRH", sep = "_"),
|
||||
col_id = NA_integer_,
|
||||
species = "group H" ,
|
||||
fullname = "Streptococcus group H",
|
||||
ref = NA_character_,
|
||||
species_id = "",
|
||||
source = "manually added"),
|
||||
MOs %>%
|
||||
filter(genus == "Streptococcus", species == "agalactiae") %>% .[1,] %>%
|
||||
mutate(mo = paste(MOs[MOs$fullname == "Streptococcus",]$mo, "GRK", sep = "_"),
|
||||
col_id = NA_integer_,
|
||||
species = "group K" ,
|
||||
fullname = "Streptococcus group K",
|
||||
ref = NA_character_,
|
||||
species_id = "",
|
||||
source = "manually added"),
|
||||
# Beta-haemolytic Streptococci
|
||||
MOs %>%
|
||||
filter(genus == "Streptococcus", species == "agalactiae") %>% .[1,] %>%
|
||||
mutate(mo = paste(MOs[MOs$fullname == "Streptococcus",]$mo, "HAE", sep = "_"),
|
||||
col_id = NA_integer_,
|
||||
species = "beta-haemolytic" ,
|
||||
fullname = "Beta-haemolytic Streptococcus",
|
||||
ref = NA_character_,
|
||||
species_id = "",
|
||||
source = "manually added")
|
||||
)
|
||||
|
||||
|
||||
# everything distinct?
|
||||
sum(duplicated(MOs$mo))
|
||||
colnames(MOs)
|
||||
|
||||
# set prevalence per species
|
||||
MOs <- MOs %>%
|
||||
mutate(prevalence = case_when(
|
||||
class == "Gammaproteobacteria"
|
||||
| genus %in% c("Enterococcus", "Staphylococcus", "Streptococcus")
|
||||
| mo %in% c("UNKNOWN", "B_GRAMN", "B_GRAMP")
|
||||
~ 1,
|
||||
phylum %in% c("Proteobacteria",
|
||||
"Firmicutes",
|
||||
"Actinobacteria",
|
||||
"Sarcomastigophora")
|
||||
| genus %in% c("Aspergillus",
|
||||
"Bacteroides",
|
||||
"Candida",
|
||||
"Capnocytophaga",
|
||||
"Chryseobacterium",
|
||||
"Cryptococcus",
|
||||
"Elisabethkingia",
|
||||
"Flavobacterium",
|
||||
"Fusobacterium",
|
||||
"Giardia",
|
||||
"Leptotrichia",
|
||||
"Mycoplasma",
|
||||
"Prevotella",
|
||||
"Rhodotorula",
|
||||
"Treponema",
|
||||
"Trichophyton",
|
||||
"Trichomonas",
|
||||
"Ureaplasma")
|
||||
| rank %in% c("kingdom", "phylum", "class", "order", "family")
|
||||
~ 2,
|
||||
TRUE ~ 3
|
||||
))
|
||||
|
||||
# arrange
|
||||
MOs <- MOs %>% arrange(fullname)
|
||||
|
||||
# transform
|
||||
MOs <- as.data.frame(MOs, stringsAsFactors = FALSE)
|
||||
MOs.old <- as.data.frame(MOs.old, stringsAsFactors = FALSE)
|
||||
class(MOs$mo) <- "mo"
|
||||
MOs$col_id <- as.integer(MOs$col_id)
|
||||
|
||||
# get differences in MO codes between this data and the package version
|
||||
MO_diff <- AMR::microorganisms %>%
|
||||
mutate(pastedtext = paste(mo, fullname)) %>%
|
||||
filter(!pastedtext %in% (MOs %>% mutate(pastedtext = paste(mo, fullname)) %>% pull(pastedtext))) %>%
|
||||
select(mo_old = mo, fullname, pastedtext) %>%
|
||||
left_join(MOs %>%
|
||||
transmute(mo_new = mo, fullname_new = fullname, pastedtext = paste(mo, fullname)), "pastedtext") %>%
|
||||
select(mo_old, mo_new, fullname_new)
|
||||
|
||||
mo_diff2 <- AMR::microorganisms %>%
|
||||
select(mo, fullname) %>%
|
||||
left_join(MOs %>%
|
||||
select(mo, fullname),
|
||||
by = "fullname",
|
||||
suffix = c("_old", "_new")) %>%
|
||||
filter(mo_old != mo_new,
|
||||
#!mo_new %in% mo_old,
|
||||
!mo_old %like% "\\[")
|
||||
|
||||
mo_diff3 <- tibble(previous_old = names(AMR:::make_trans_tbl()),
|
||||
previous_new = AMR:::make_trans_tbl()) %>%
|
||||
left_join(AMR::microorganisms %>% select(mo, fullname), by = c(previous_new = "mo")) %>%
|
||||
left_join(MOs %>% select(mo_new = mo, fullname), by = "fullname")
|
||||
|
||||
# what did we win most?
|
||||
MOs %>% filter(!fullname %in% AMR::microorganisms$fullname) %>% freq(genus)
|
||||
# what did we lose most?
|
||||
AMR::microorganisms %>%
|
||||
filter(kingdom != "Chromista" & !fullname %in% MOs$fullname & !fullname %in% MOs.old$fullname) %>%
|
||||
freq(genus)
|
||||
|
||||
|
||||
# save
|
||||
saveRDS(MOs, "microorganisms.rds")
|
||||
saveRDS(MOs.old, "microorganisms.old.rds")
|
||||
|
||||
# on the server, do:
|
||||
usethis::use_data(microorganisms, overwrite = TRUE, version = 2)
|
||||
usethis::use_data(microorganisms.old, overwrite = TRUE, version = 2)
|
||||
rm(microorganisms)
|
||||
rm(microorganisms.old)
|
||||
# and update the year in R/data.R
|
||||
@ -78,7 +78,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9008</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9023</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -156,13 +156,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
|
Before 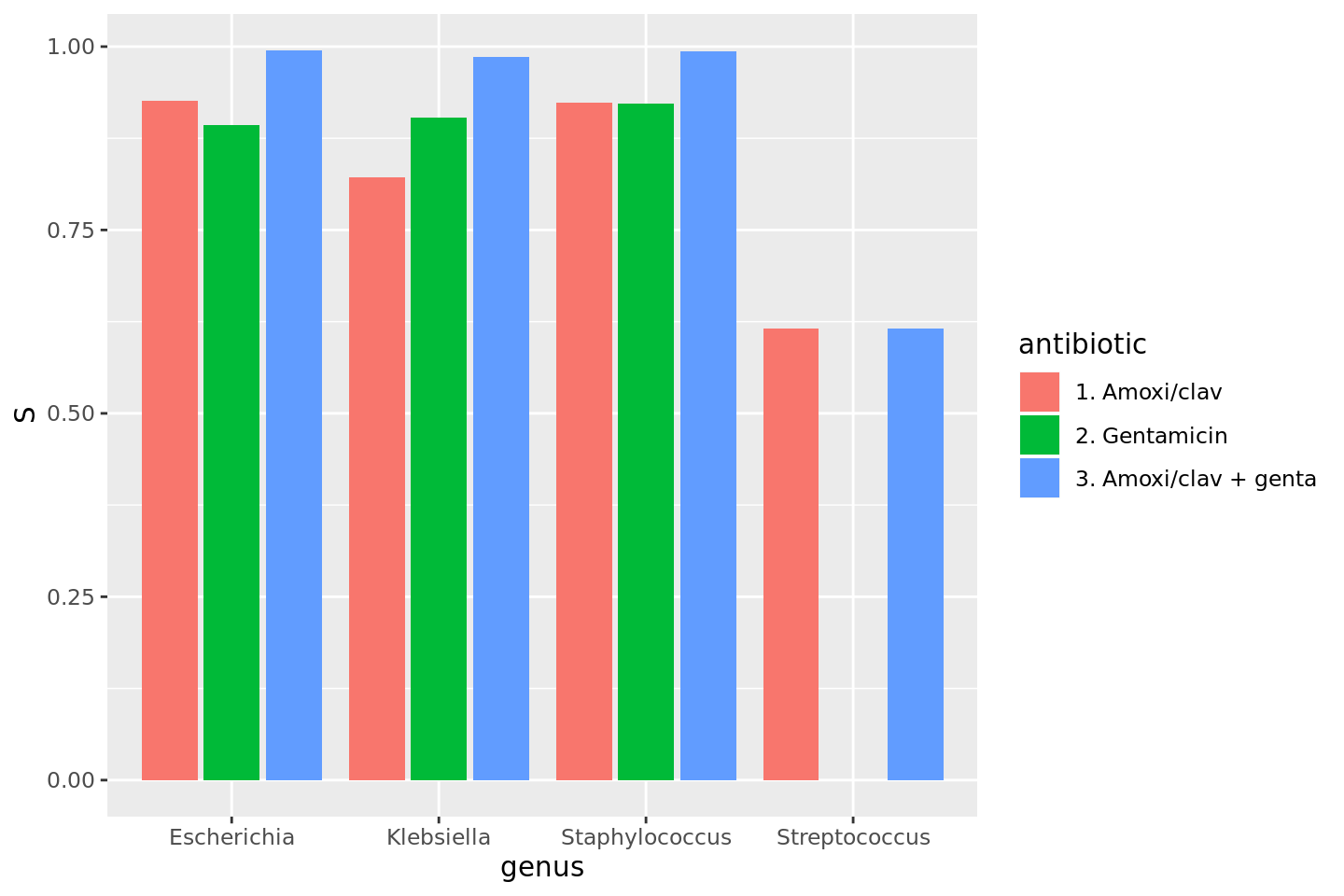
(image error) Size: 35 KiB After 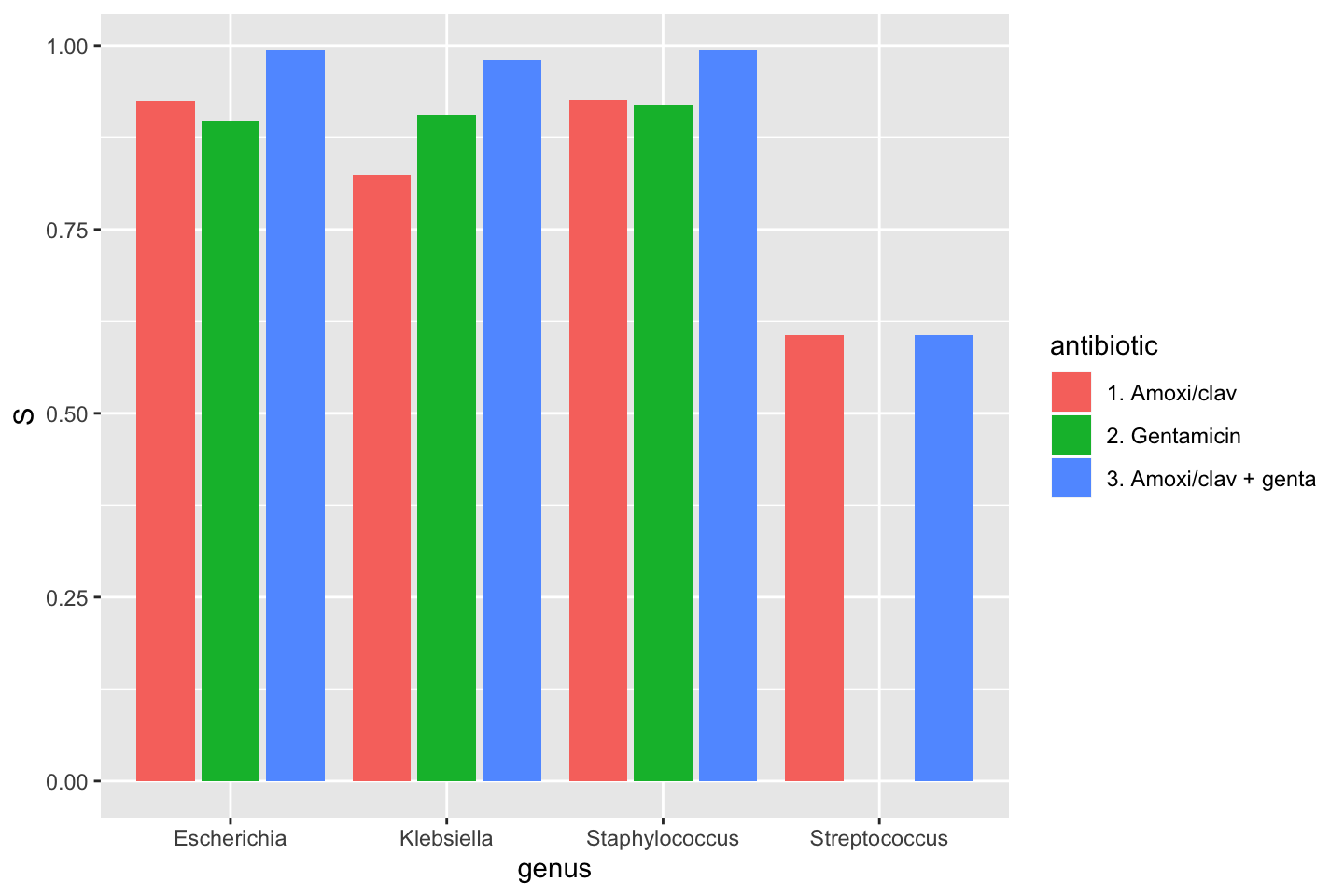
(image error) Size: 63 KiB 

|
|
Before 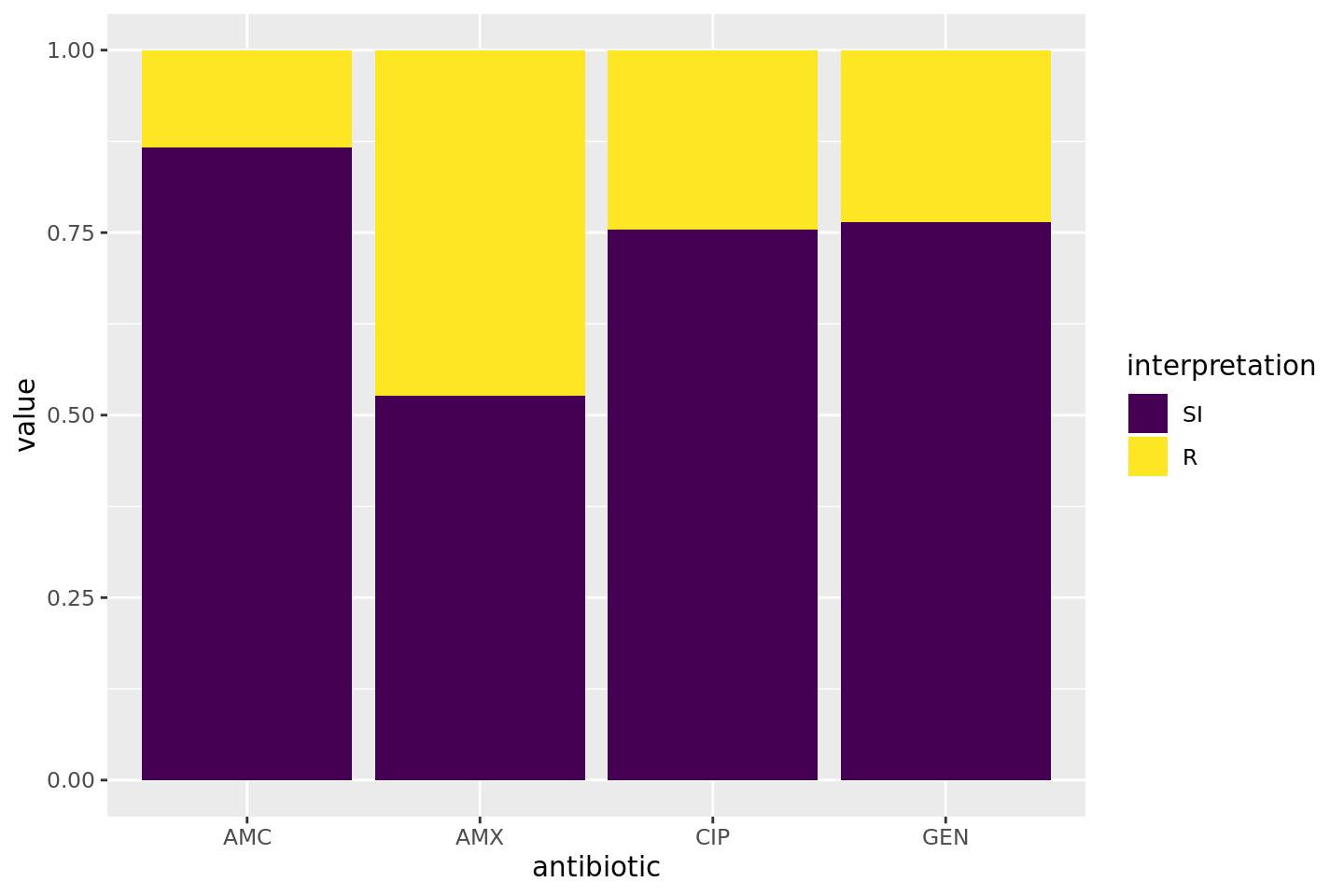
(image error) Size: 18 KiB After 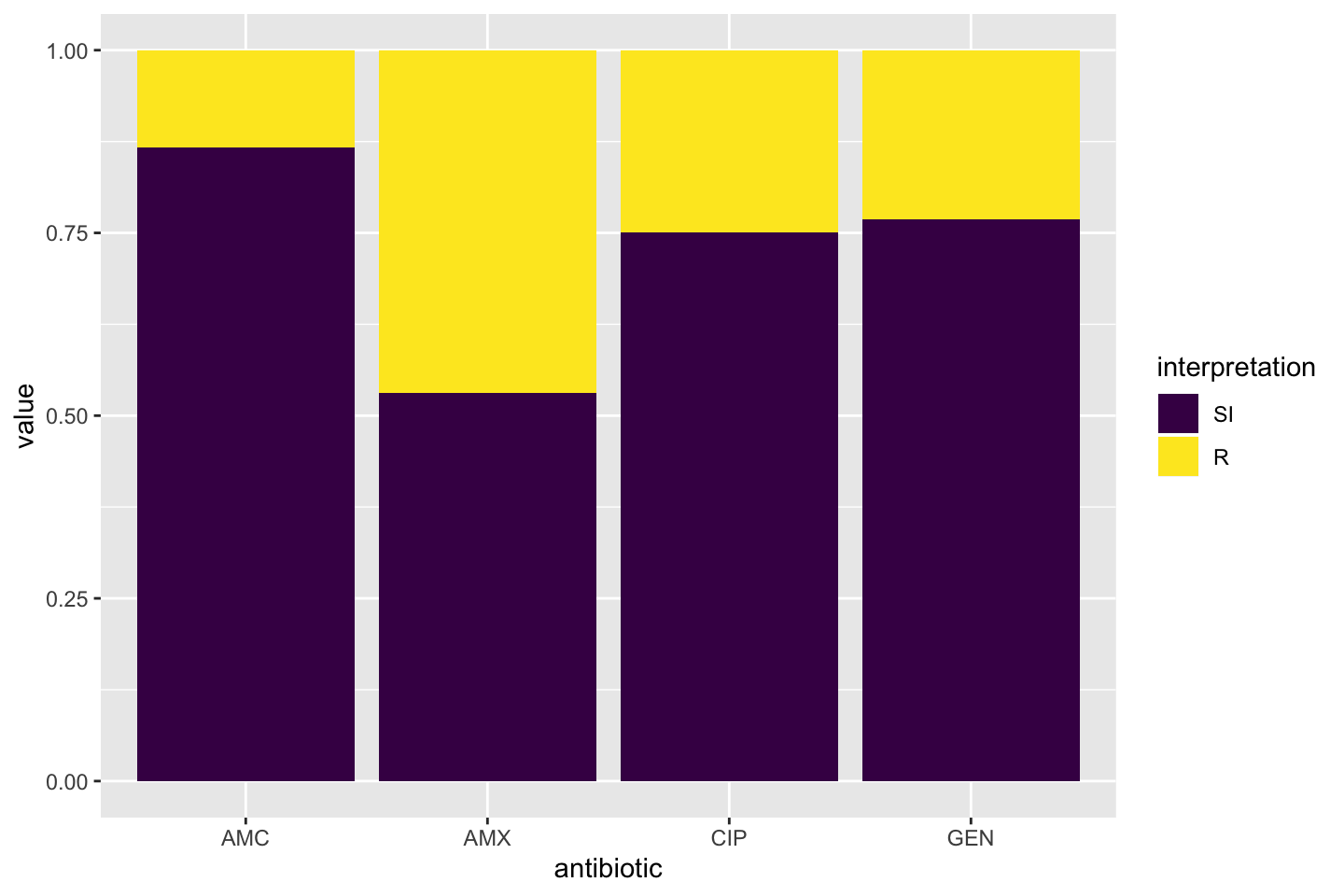
(image error) Size: 51 KiB 

|
|
Before 
(image error) Size: 68 KiB After 
(image error) Size: 102 KiB 

|
|
Before 
(image error) Size: 42 KiB After 
(image error) Size: 77 KiB 

|
@ -40,7 +40,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -118,13 +118,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -192,7 +185,7 @@
|
||||
<h1>How to apply EUCAST rules</h1>
|
||||
<h4 class="author">Matthijs S. Berends</h4>
|
||||
|
||||
<h4 class="date">01 July 2019</h4>
|
||||
<h4 class="date">29 July 2019</h4>
|
||||
|
||||
|
||||
<div class="hidden name"><code>EUCAST.Rmd</code></div>
|
||||
|
||||
@ -40,7 +40,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9026</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -118,13 +118,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -192,7 +185,7 @@
|
||||
<h1>How to determine multi-drug resistance (MDR)</h1>
|
||||
<h4 class="author">Matthijs S. Berends</h4>
|
||||
|
||||
<h4 class="date">01 July 2019</h4>
|
||||
<h4 class="date">06 August 2019</h4>
|
||||
|
||||
|
||||
<div class="hidden name"><code>MDR.Rmd</code></div>
|
||||
@ -208,58 +201,60 @@
|
||||
<li>“WIP-Richtlijn Bijzonder Resistente Micro-organismen (BRMO)”, by RIVM (Rijksinstituut voor de Volksgezondheid, the Netherlands National Institute for Public Health and the Environment)</li>
|
||||
</ul>
|
||||
<p>As an example, I will make a data set to determine multi-drug resistant TB:</p>
|
||||
<div class="sourceCode" id="cb1"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb1-1" title="1"><span class="co"># a helper function to get a random vector with values S, I and R</span></a>
|
||||
<a class="sourceLine" id="cb1-2" title="2"><span class="co"># with the probabilities 50%-10%-40%</span></a>
|
||||
<a class="sourceLine" id="cb1-3" title="3">sample_rsi <-<span class="st"> </span><span class="cf">function</span>() {</a>
|
||||
<a class="sourceLine" id="cb1-4" title="4"> <span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/sample">sample</a></span>(<span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/c">c</a></span>(<span class="st">"S"</span>, <span class="st">"I"</span>, <span class="st">"R"</span>),</a>
|
||||
<a class="sourceLine" id="cb1-5" title="5"> <span class="dt">size =</span> <span class="dv">5000</span>,</a>
|
||||
<a class="sourceLine" id="cb1-6" title="6"> <span class="dt">prob =</span> <span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/c">c</a></span>(<span class="fl">0.5</span>, <span class="fl">0.1</span>, <span class="fl">0.4</span>),</a>
|
||||
<a class="sourceLine" id="cb1-7" title="7"> <span class="dt">replace =</span> <span class="ot">TRUE</span>)</a>
|
||||
<a class="sourceLine" id="cb1-8" title="8">}</a>
|
||||
<a class="sourceLine" id="cb1-9" title="9"></a>
|
||||
<a class="sourceLine" id="cb1-10" title="10">my_TB_data <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/data.frame">data.frame</a></span>(<span class="dt">rifampicin =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb1-11" title="11"> <span class="dt">isoniazid =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb1-12" title="12"> <span class="dt">gatifloxacin =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb1-13" title="13"> <span class="dt">ethambutol =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb1-14" title="14"> <span class="dt">pyrazinamide =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb1-15" title="15"> <span class="dt">moxifloxacin =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb1-16" title="16"> <span class="dt">kanamycin =</span> <span class="kw">sample_rsi</span>())</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb1"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb1-1" data-line-number="1"><span class="co"># a helper function to get a random vector with values S, I and R</span></a>
|
||||
<a class="sourceLine" id="cb1-2" data-line-number="2"><span class="co"># with the probabilities 50%-10%-40%</span></a>
|
||||
<a class="sourceLine" id="cb1-3" data-line-number="3">sample_rsi <-<span class="st"> </span><span class="cf">function</span>() {</a>
|
||||
<a class="sourceLine" id="cb1-4" data-line-number="4"> <span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/sample">sample</a></span>(<span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/c">c</a></span>(<span class="st">"S"</span>, <span class="st">"I"</span>, <span class="st">"R"</span>),</a>
|
||||
<a class="sourceLine" id="cb1-5" data-line-number="5"> <span class="dt">size =</span> <span class="dv">5000</span>,</a>
|
||||
<a class="sourceLine" id="cb1-6" data-line-number="6"> <span class="dt">prob =</span> <span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/c">c</a></span>(<span class="fl">0.5</span>, <span class="fl">0.1</span>, <span class="fl">0.4</span>),</a>
|
||||
<a class="sourceLine" id="cb1-7" data-line-number="7"> <span class="dt">replace =</span> <span class="ot">TRUE</span>)</a>
|
||||
<a class="sourceLine" id="cb1-8" data-line-number="8">}</a>
|
||||
<a class="sourceLine" id="cb1-9" data-line-number="9"></a>
|
||||
<a class="sourceLine" id="cb1-10" data-line-number="10">my_TB_data <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/data.frame">data.frame</a></span>(<span class="dt">rifampicin =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb1-11" data-line-number="11"> <span class="dt">isoniazid =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb1-12" data-line-number="12"> <span class="dt">gatifloxacin =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb1-13" data-line-number="13"> <span class="dt">ethambutol =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb1-14" data-line-number="14"> <span class="dt">pyrazinamide =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb1-15" data-line-number="15"> <span class="dt">moxifloxacin =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb1-16" data-line-number="16"> <span class="dt">kanamycin =</span> <span class="kw">sample_rsi</span>())</a></code></pre></div>
|
||||
<p>Because all column names are automatically verified for valid drug names or codes, this would have worked exactly the same:</p>
|
||||
<div class="sourceCode" id="cb2"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb2-1" title="1">my_TB_data <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/data.frame">data.frame</a></span>(<span class="dt">RIF =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb2-2" title="2"> <span class="dt">INH =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb2-3" title="3"> <span class="dt">GAT =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb2-4" title="4"> <span class="dt">ETH =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb2-5" title="5"> <span class="dt">PZA =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb2-6" title="6"> <span class="dt">MFX =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb2-7" title="7"> <span class="dt">KAN =</span> <span class="kw">sample_rsi</span>())</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb2"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb2-1" data-line-number="1">my_TB_data <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/data.frame">data.frame</a></span>(<span class="dt">RIF =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb2-2" data-line-number="2"> <span class="dt">INH =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb2-3" data-line-number="3"> <span class="dt">GAT =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb2-4" data-line-number="4"> <span class="dt">ETH =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb2-5" data-line-number="5"> <span class="dt">PZA =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb2-6" data-line-number="6"> <span class="dt">MFX =</span> <span class="kw">sample_rsi</span>(),</a>
|
||||
<a class="sourceLine" id="cb2-7" data-line-number="7"> <span class="dt">KAN =</span> <span class="kw">sample_rsi</span>())</a></code></pre></div>
|
||||
<p>The data set looks like this now:</p>
|
||||
<div class="sourceCode" id="cb3"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb3-1" title="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/utils/topics/head">head</a></span>(my_TB_data)</a>
|
||||
<a class="sourceLine" id="cb3-2" title="2"><span class="co"># rifampicin isoniazid gatifloxacin ethambutol pyrazinamide moxifloxacin</span></a>
|
||||
<a class="sourceLine" id="cb3-3" title="3"><span class="co"># 1 R R S S R I</span></a>
|
||||
<a class="sourceLine" id="cb3-4" title="4"><span class="co"># 2 R R S R S R</span></a>
|
||||
<a class="sourceLine" id="cb3-5" title="5"><span class="co"># 3 R S S R R R</span></a>
|
||||
<a class="sourceLine" id="cb3-6" title="6"><span class="co"># 4 R R S S S I</span></a>
|
||||
<a class="sourceLine" id="cb3-7" title="7"><span class="co"># 5 R R S S R I</span></a>
|
||||
<a class="sourceLine" id="cb3-8" title="8"><span class="co"># 6 R S R R S S</span></a>
|
||||
<a class="sourceLine" id="cb3-9" title="9"><span class="co"># kanamycin</span></a>
|
||||
<a class="sourceLine" id="cb3-10" title="10"><span class="co"># 1 S</span></a>
|
||||
<a class="sourceLine" id="cb3-11" title="11"><span class="co"># 2 S</span></a>
|
||||
<a class="sourceLine" id="cb3-12" title="12"><span class="co"># 3 S</span></a>
|
||||
<a class="sourceLine" id="cb3-13" title="13"><span class="co"># 4 R</span></a>
|
||||
<a class="sourceLine" id="cb3-14" title="14"><span class="co"># 5 R</span></a>
|
||||
<a class="sourceLine" id="cb3-15" title="15"><span class="co"># 6 S</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb3"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb3-1" data-line-number="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/utils/topics/head">head</a></span>(my_TB_data)</a>
|
||||
<a class="sourceLine" id="cb3-2" data-line-number="2"><span class="co"># rifampicin isoniazid gatifloxacin ethambutol pyrazinamide moxifloxacin</span></a>
|
||||
<a class="sourceLine" id="cb3-3" data-line-number="3"><span class="co"># 1 R R S R S S</span></a>
|
||||
<a class="sourceLine" id="cb3-4" data-line-number="4"><span class="co"># 2 R R S R S S</span></a>
|
||||
<a class="sourceLine" id="cb3-5" data-line-number="5"><span class="co"># 3 R S R S S R</span></a>
|
||||
<a class="sourceLine" id="cb3-6" data-line-number="6"><span class="co"># 4 R I S R R R</span></a>
|
||||
<a class="sourceLine" id="cb3-7" data-line-number="7"><span class="co"># 5 I S S R I S</span></a>
|
||||
<a class="sourceLine" id="cb3-8" data-line-number="8"><span class="co"># 6 R R R S I S</span></a>
|
||||
<a class="sourceLine" id="cb3-9" data-line-number="9"><span class="co"># kanamycin</span></a>
|
||||
<a class="sourceLine" id="cb3-10" data-line-number="10"><span class="co"># 1 I</span></a>
|
||||
<a class="sourceLine" id="cb3-11" data-line-number="11"><span class="co"># 2 S</span></a>
|
||||
<a class="sourceLine" id="cb3-12" data-line-number="12"><span class="co"># 3 I</span></a>
|
||||
<a class="sourceLine" id="cb3-13" data-line-number="13"><span class="co"># 4 S</span></a>
|
||||
<a class="sourceLine" id="cb3-14" data-line-number="14"><span class="co"># 5 S</span></a>
|
||||
<a class="sourceLine" id="cb3-15" data-line-number="15"><span class="co"># 6 S</span></a></code></pre></div>
|
||||
<p>We can now add the interpretation of MDR-TB to our data set:</p>
|
||||
<div class="sourceCode" id="cb4"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb4-1" title="1">my_TB_data<span class="op">$</span>mdr <-<span class="st"> </span><span class="kw"><a href="../reference/mdro.html">mdr_tb</a></span>(my_TB_data)</a>
|
||||
<a class="sourceLine" id="cb4-2" title="2"><span class="co"># </span><span class="al">NOTE</span><span class="co">: No column found as input for `col_mo`, assuming all records contain Mycobacterium tuberculosis.</span></a>
|
||||
<a class="sourceLine" id="cb4-3" title="3"><span class="co"># Determining multidrug-resistant organisms (MDRO), according to:</span></a>
|
||||
<a class="sourceLine" id="cb4-4" title="4"><span class="co"># Guideline: Companion handbook to the WHO guidelines for the programmatic management of drug-resistant tuberculosis</span></a>
|
||||
<a class="sourceLine" id="cb4-5" title="5"><span class="co"># Version: WHO/HTM/TB/2014.11</span></a>
|
||||
<a class="sourceLine" id="cb4-6" title="6"><span class="co"># Author: WHO (World Health Organization)</span></a>
|
||||
<a class="sourceLine" id="cb4-7" title="7"><span class="co"># Source: https://www.who.int/tb/publications/pmdt_companionhandbook/en/</span></a>
|
||||
<a class="sourceLine" id="cb4-8" title="8"><span class="co"># </span><span class="al">NOTE</span><span class="co">: Reliability might be improved if these antimicrobial results would be available too: CAP (capreomycin), RIB (rifabutin), RFP (rifapentine)</span></a></code></pre></div>
|
||||
<p>And review the result with a frequency table:</p>
|
||||
<div class="sourceCode" id="cb5"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb5-1" title="1"><span class="kw"><a href="../reference/freq.html">freq</a></span>(my_TB_data<span class="op">$</span>mdr)</a></code></pre></div>
|
||||
<p><strong>Frequency table of <code>mdr</code> from <code>my_TB_data</code> (5,000 x 8)</strong></p>
|
||||
<div class="sourceCode" id="cb4"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb4-1" data-line-number="1">my_TB_data<span class="op">$</span>mdr <-<span class="st"> </span><span class="kw"><a href="../reference/mdro.html">mdr_tb</a></span>(my_TB_data)</a>
|
||||
<a class="sourceLine" id="cb4-2" data-line-number="2"><span class="co"># </span><span class="al">NOTE</span><span class="co">: No column found as input for `col_mo`, assuming all records contain Mycobacterium tuberculosis.</span></a>
|
||||
<a class="sourceLine" id="cb4-3" data-line-number="3"><span class="co"># Determining multidrug-resistant organisms (MDRO), according to:</span></a>
|
||||
<a class="sourceLine" id="cb4-4" data-line-number="4"><span class="co"># Guideline: Companion handbook to the WHO guidelines for the programmatic management of drug-resistant tuberculosis</span></a>
|
||||
<a class="sourceLine" id="cb4-5" data-line-number="5"><span class="co"># Version: WHO/HTM/TB/2014.11</span></a>
|
||||
<a class="sourceLine" id="cb4-6" data-line-number="6"><span class="co"># Author: WHO (World Health Organization)</span></a>
|
||||
<a class="sourceLine" id="cb4-7" data-line-number="7"><span class="co"># Source: https://www.who.int/tb/publications/pmdt_companionhandbook/en/</span></a>
|
||||
<a class="sourceLine" id="cb4-8" data-line-number="8"><span class="co"># </span><span class="al">NOTE</span><span class="co">: Reliability might be improved if these antimicrobial results would be available too: CAP (capreomycin), RIB (rifabutin), RFP (rifapentine)</span></a></code></pre></div>
|
||||
<p>We also created a package dedicated to data cleaning and checking, called the <code>clean</code> package. It gets automatically installed with the <code>AMR</code> package, so we only have to load it:</p>
|
||||
<div class="sourceCode" id="cb5"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb5-1" data-line-number="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(clean)</a></code></pre></div>
|
||||
<p>It contains the <code><a href="https://www.rdocumentation.org/packages/clean/topics/freq">freq()</a></code> function, to create a frequency table:</p>
|
||||
<div class="sourceCode" id="cb6"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb6-1" data-line-number="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/clean/topics/freq">freq</a></span>(my_TB_data<span class="op">$</span>mdr)</a></code></pre></div>
|
||||
<p><strong>Frequency table</strong></p>
|
||||
<p>Class: factor > ordered (numeric)<br>
|
||||
Length: 5,000 (of which NA: 0 = 0.00%)<br>
|
||||
Levels: 5: Negative < Mono-resistance < Poly-resistance < Multidrug resistance…<br>
|
||||
@ -277,41 +272,41 @@ Unique: 5</p>
|
||||
<tr class="odd">
|
||||
<td align="left">1</td>
|
||||
<td align="left">Mono-resistance</td>
|
||||
<td align="right">3,222</td>
|
||||
<td align="right">64.4%</td>
|
||||
<td align="right">3,222</td>
|
||||
<td align="right">64.4%</td>
|
||||
<td align="right">3264</td>
|
||||
<td align="right">65.3%</td>
|
||||
<td align="right">3264</td>
|
||||
<td align="right">65.3%</td>
|
||||
</tr>
|
||||
<tr class="even">
|
||||
<td align="left">2</td>
|
||||
<td align="left">Negative</td>
|
||||
<td align="right">659</td>
|
||||
<td align="right">13.2%</td>
|
||||
<td align="right">3,881</td>
|
||||
<td align="right">77.6%</td>
|
||||
<td align="right">627</td>
|
||||
<td align="right">12.5%</td>
|
||||
<td align="right">3891</td>
|
||||
<td align="right">77.8%</td>
|
||||
</tr>
|
||||
<tr class="odd">
|
||||
<td align="left">3</td>
|
||||
<td align="left">Multidrug resistance</td>
|
||||
<td align="right">589</td>
|
||||
<td align="right">11.8%</td>
|
||||
<td align="right">4,470</td>
|
||||
<td align="right">89.4%</td>
|
||||
<td align="right">607</td>
|
||||
<td align="right">12.1%</td>
|
||||
<td align="right">4498</td>
|
||||
<td align="right">90.0%</td>
|
||||
</tr>
|
||||
<tr class="even">
|
||||
<td align="left">4</td>
|
||||
<td align="left">Poly-resistance</td>
|
||||
<td align="right">313</td>
|
||||
<td align="right">6.3%</td>
|
||||
<td align="right">4,783</td>
|
||||
<td align="right">288</td>
|
||||
<td align="right">5.8%</td>
|
||||
<td align="right">4786</td>
|
||||
<td align="right">95.7%</td>
|
||||
</tr>
|
||||
<tr class="odd">
|
||||
<td align="left">5</td>
|
||||
<td align="left">Extensive drug resistance</td>
|
||||
<td align="right">217</td>
|
||||
<td align="right">214</td>
|
||||
<td align="right">4.3%</td>
|
||||
<td align="right">5,000</td>
|
||||
<td align="right">5000</td>
|
||||
<td align="right">100.0%</td>
|
||||
</tr>
|
||||
</tbody>
|
||||
|
||||
@ -40,7 +40,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -118,13 +118,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -192,7 +185,7 @@
|
||||
<h1>How to import data from SPSS / SAS / Stata</h1>
|
||||
<h4 class="author">Matthijs S. Berends</h4>
|
||||
|
||||
<h4 class="date">01 July 2019</h4>
|
||||
<h4 class="date">29 July 2019</h4>
|
||||
|
||||
|
||||
<div class="hidden name"><code>SPSS.Rmd</code></div>
|
||||
@ -242,39 +235,39 @@
|
||||
</ul>
|
||||
<p>If you sometimes write syntaxes in SPSS to run a complete analysis or to ‘automate’ some of your work, you should perhaps do this in R. You will notice that writing syntaxes in R is a lot more nifty and clever than in SPSS. Still, as working with any statistical package, you will have to have knowledge about what you are doing (statistically) and what you are willing to accomplish.</p>
|
||||
<p>To demonstrate the first point:</p>
|
||||
<div class="sourceCode" id="cb1"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb1-1" title="1"><span class="co"># not all values are valid MIC values:</span></a>
|
||||
<a class="sourceLine" id="cb1-2" title="2"><span class="kw"><a href="../reference/as.mic.html">as.mic</a></span>(<span class="fl">0.125</span>)</a>
|
||||
<a class="sourceLine" id="cb1-3" title="3"><span class="co"># Class 'mic'</span></a>
|
||||
<a class="sourceLine" id="cb1-4" title="4"><span class="co"># [1] 0.125</span></a>
|
||||
<a class="sourceLine" id="cb1-5" title="5"><span class="kw"><a href="../reference/as.mic.html">as.mic</a></span>(<span class="st">"testvalue"</span>)</a>
|
||||
<a class="sourceLine" id="cb1-6" title="6"><span class="co"># Class 'mic'</span></a>
|
||||
<a class="sourceLine" id="cb1-7" title="7"><span class="co"># [1] <NA></span></a>
|
||||
<a class="sourceLine" id="cb1-8" title="8"></a>
|
||||
<a class="sourceLine" id="cb1-9" title="9"><span class="co"># the Gram stain is avaiable for all bacteria:</span></a>
|
||||
<a class="sourceLine" id="cb1-10" title="10"><span class="kw"><a href="../reference/mo_property.html">mo_gramstain</a></span>(<span class="st">"E. coli"</span>)</a>
|
||||
<a class="sourceLine" id="cb1-11" title="11"><span class="co"># [1] "Gram-negative"</span></a>
|
||||
<a class="sourceLine" id="cb1-12" title="12"></a>
|
||||
<a class="sourceLine" id="cb1-13" title="13"><span class="co"># Klebsiella is intrinsic resistant to amoxicllin, according to EUCAST:</span></a>
|
||||
<a class="sourceLine" id="cb1-14" title="14">klebsiella_test <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/data.frame">data.frame</a></span>(<span class="dt">mo =</span> <span class="st">"klebsiella"</span>, </a>
|
||||
<a class="sourceLine" id="cb1-15" title="15"> <span class="dt">amox =</span> <span class="st">"S"</span>,</a>
|
||||
<a class="sourceLine" id="cb1-16" title="16"> <span class="dt">stringsAsFactors =</span> <span class="ot">FALSE</span>)</a>
|
||||
<a class="sourceLine" id="cb1-17" title="17">klebsiella_test</a>
|
||||
<a class="sourceLine" id="cb1-18" title="18"><span class="co"># mo amox</span></a>
|
||||
<a class="sourceLine" id="cb1-19" title="19"><span class="co"># 1 klebsiella S</span></a>
|
||||
<a class="sourceLine" id="cb1-20" title="20"><span class="kw"><a href="../reference/eucast_rules.html">eucast_rules</a></span>(klebsiella_test, <span class="dt">info =</span> <span class="ot">FALSE</span>)</a>
|
||||
<a class="sourceLine" id="cb1-21" title="21"><span class="co"># mo amox</span></a>
|
||||
<a class="sourceLine" id="cb1-22" title="22"><span class="co"># 1 klebsiella R</span></a>
|
||||
<a class="sourceLine" id="cb1-23" title="23"></a>
|
||||
<a class="sourceLine" id="cb1-24" title="24"><span class="co"># hundreds of trade names can be translated to a name, trade name or an ATC code:</span></a>
|
||||
<a class="sourceLine" id="cb1-25" title="25"><span class="kw"><a href="../reference/ab_property.html">ab_name</a></span>(<span class="st">"floxapen"</span>)</a>
|
||||
<a class="sourceLine" id="cb1-26" title="26"><span class="co"># [1] "Flucloxacillin"</span></a>
|
||||
<a class="sourceLine" id="cb1-27" title="27"><span class="kw"><a href="../reference/ab_property.html">ab_tradenames</a></span>(<span class="st">"floxapen"</span>)</a>
|
||||
<a class="sourceLine" id="cb1-28" title="28"><span class="co"># [1] "Floxacillin" "FLOXACILLIN" "Floxapen" </span></a>
|
||||
<a class="sourceLine" id="cb1-29" title="29"><span class="co"># [4] "Floxapen sodium salt" "Fluclox" "Flucloxacilina" </span></a>
|
||||
<a class="sourceLine" id="cb1-30" title="30"><span class="co"># [7] "Flucloxacillin" "Flucloxacilline" "Flucloxacillinum" </span></a>
|
||||
<a class="sourceLine" id="cb1-31" title="31"><span class="co"># [10] "Fluorochloroxacillin"</span></a>
|
||||
<a class="sourceLine" id="cb1-32" title="32"><span class="kw"><a href="../reference/ab_property.html">ab_atc</a></span>(<span class="st">"floxapen"</span>)</a>
|
||||
<a class="sourceLine" id="cb1-33" title="33"><span class="co"># [1] "J01CF05"</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb1"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb1-1" data-line-number="1"><span class="co"># not all values are valid MIC values:</span></a>
|
||||
<a class="sourceLine" id="cb1-2" data-line-number="2"><span class="kw"><a href="../reference/as.mic.html">as.mic</a></span>(<span class="fl">0.125</span>)</a>
|
||||
<a class="sourceLine" id="cb1-3" data-line-number="3"><span class="co"># Class 'mic'</span></a>
|
||||
<a class="sourceLine" id="cb1-4" data-line-number="4"><span class="co"># [1] 0.125</span></a>
|
||||
<a class="sourceLine" id="cb1-5" data-line-number="5"><span class="kw"><a href="../reference/as.mic.html">as.mic</a></span>(<span class="st">"testvalue"</span>)</a>
|
||||
<a class="sourceLine" id="cb1-6" data-line-number="6"><span class="co"># Class 'mic'</span></a>
|
||||
<a class="sourceLine" id="cb1-7" data-line-number="7"><span class="co"># [1] <NA></span></a>
|
||||
<a class="sourceLine" id="cb1-8" data-line-number="8"></a>
|
||||
<a class="sourceLine" id="cb1-9" data-line-number="9"><span class="co"># the Gram stain is avaiable for all bacteria:</span></a>
|
||||
<a class="sourceLine" id="cb1-10" data-line-number="10"><span class="kw"><a href="../reference/mo_property.html">mo_gramstain</a></span>(<span class="st">"E. coli"</span>)</a>
|
||||
<a class="sourceLine" id="cb1-11" data-line-number="11"><span class="co"># [1] "Gram-negative"</span></a>
|
||||
<a class="sourceLine" id="cb1-12" data-line-number="12"></a>
|
||||
<a class="sourceLine" id="cb1-13" data-line-number="13"><span class="co"># Klebsiella is intrinsic resistant to amoxicllin, according to EUCAST:</span></a>
|
||||
<a class="sourceLine" id="cb1-14" data-line-number="14">klebsiella_test <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/data.frame">data.frame</a></span>(<span class="dt">mo =</span> <span class="st">"klebsiella"</span>, </a>
|
||||
<a class="sourceLine" id="cb1-15" data-line-number="15"> <span class="dt">amox =</span> <span class="st">"S"</span>,</a>
|
||||
<a class="sourceLine" id="cb1-16" data-line-number="16"> <span class="dt">stringsAsFactors =</span> <span class="ot">FALSE</span>)</a>
|
||||
<a class="sourceLine" id="cb1-17" data-line-number="17">klebsiella_test</a>
|
||||
<a class="sourceLine" id="cb1-18" data-line-number="18"><span class="co"># mo amox</span></a>
|
||||
<a class="sourceLine" id="cb1-19" data-line-number="19"><span class="co"># 1 klebsiella S</span></a>
|
||||
<a class="sourceLine" id="cb1-20" data-line-number="20"><span class="kw"><a href="../reference/eucast_rules.html">eucast_rules</a></span>(klebsiella_test, <span class="dt">info =</span> <span class="ot">FALSE</span>)</a>
|
||||
<a class="sourceLine" id="cb1-21" data-line-number="21"><span class="co"># mo amox</span></a>
|
||||
<a class="sourceLine" id="cb1-22" data-line-number="22"><span class="co"># 1 klebsiella R</span></a>
|
||||
<a class="sourceLine" id="cb1-23" data-line-number="23"></a>
|
||||
<a class="sourceLine" id="cb1-24" data-line-number="24"><span class="co"># hundreds of trade names can be translated to a name, trade name or an ATC code:</span></a>
|
||||
<a class="sourceLine" id="cb1-25" data-line-number="25"><span class="kw"><a href="../reference/ab_property.html">ab_name</a></span>(<span class="st">"floxapen"</span>)</a>
|
||||
<a class="sourceLine" id="cb1-26" data-line-number="26"><span class="co"># [1] "Flucloxacillin"</span></a>
|
||||
<a class="sourceLine" id="cb1-27" data-line-number="27"><span class="kw"><a href="../reference/ab_property.html">ab_tradenames</a></span>(<span class="st">"floxapen"</span>)</a>
|
||||
<a class="sourceLine" id="cb1-28" data-line-number="28"><span class="co"># [1] "Floxacillin" "FLOXACILLIN" "Floxapen" </span></a>
|
||||
<a class="sourceLine" id="cb1-29" data-line-number="29"><span class="co"># [4] "Floxapen sodium salt" "Fluclox" "Flucloxacilina" </span></a>
|
||||
<a class="sourceLine" id="cb1-30" data-line-number="30"><span class="co"># [7] "Flucloxacillin" "Flucloxacilline" "Flucloxacillinum" </span></a>
|
||||
<a class="sourceLine" id="cb1-31" data-line-number="31"><span class="co"># [10] "Fluorochloroxacillin"</span></a>
|
||||
<a class="sourceLine" id="cb1-32" data-line-number="32"><span class="kw"><a href="../reference/ab_property.html">ab_atc</a></span>(<span class="st">"floxapen"</span>)</a>
|
||||
<a class="sourceLine" id="cb1-33" data-line-number="33"><span class="co"># [1] "J01CF05"</span></a></code></pre></div>
|
||||
</div>
|
||||
<div id="import-data-from-spsssasstata" class="section level2">
|
||||
<h2 class="hasAnchor">
|
||||
@ -290,97 +283,97 @@
|
||||
<p><img src="https://gitlab.com/msberends/AMR/raw/master/docs/import2.png"></p>
|
||||
<p>If you want named variables to be imported as factors so it resembles SPSS more, use <code><a href="https://haven.tidyverse.org/reference/as_factor.html">as_factor()</a></code>.</p>
|
||||
<p>The difference is this:</p>
|
||||
<div class="sourceCode" id="cb2"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb2-1" title="1">SPSS_data</a>
|
||||
<a class="sourceLine" id="cb2-2" title="2"><span class="co"># # A tibble: 4,203 x 4</span></a>
|
||||
<a class="sourceLine" id="cb2-3" title="3"><span class="co"># v001 sex status statusage</span></a>
|
||||
<a class="sourceLine" id="cb2-4" title="4"><span class="co"># <dbl> <dbl+lbl> <dbl+lbl> <dbl></span></a>
|
||||
<a class="sourceLine" id="cb2-5" title="5"><span class="co"># 1 10002 1 1 76.6</span></a>
|
||||
<a class="sourceLine" id="cb2-6" title="6"><span class="co"># 2 10004 0 1 59.1</span></a>
|
||||
<a class="sourceLine" id="cb2-7" title="7"><span class="co"># 3 10005 1 1 54.5</span></a>
|
||||
<a class="sourceLine" id="cb2-8" title="8"><span class="co"># 4 10006 1 1 54.1</span></a>
|
||||
<a class="sourceLine" id="cb2-9" title="9"><span class="co"># 5 10007 1 1 57.7</span></a>
|
||||
<a class="sourceLine" id="cb2-10" title="10"><span class="co"># 6 10008 1 1 62.8</span></a>
|
||||
<a class="sourceLine" id="cb2-11" title="11"><span class="co"># 7 10010 0 1 63.7</span></a>
|
||||
<a class="sourceLine" id="cb2-12" title="12"><span class="co"># 8 10011 1 1 73.1</span></a>
|
||||
<a class="sourceLine" id="cb2-13" title="13"><span class="co"># 9 10017 1 1 56.7</span></a>
|
||||
<a class="sourceLine" id="cb2-14" title="14"><span class="co"># 10 10018 0 1 66.6</span></a>
|
||||
<a class="sourceLine" id="cb2-15" title="15"><span class="co"># # … with 4,193 more rows</span></a>
|
||||
<a class="sourceLine" id="cb2-16" title="16"></a>
|
||||
<a class="sourceLine" id="cb2-17" title="17"><span class="kw">as_factor</span>(SPSS_data)</a>
|
||||
<a class="sourceLine" id="cb2-18" title="18"><span class="co"># # A tibble: 4,203 x 4</span></a>
|
||||
<a class="sourceLine" id="cb2-19" title="19"><span class="co"># v001 sex status statusage</span></a>
|
||||
<a class="sourceLine" id="cb2-20" title="20"><span class="co"># <dbl> <fct> <fct> <dbl></span></a>
|
||||
<a class="sourceLine" id="cb2-21" title="21"><span class="co"># 1 10002 Male alive 76.6</span></a>
|
||||
<a class="sourceLine" id="cb2-22" title="22"><span class="co"># 2 10004 Female alive 59.1</span></a>
|
||||
<a class="sourceLine" id="cb2-23" title="23"><span class="co"># 3 10005 Male alive 54.5</span></a>
|
||||
<a class="sourceLine" id="cb2-24" title="24"><span class="co"># 4 10006 Male alive 54.1</span></a>
|
||||
<a class="sourceLine" id="cb2-25" title="25"><span class="co"># 5 10007 Male alive 57.7</span></a>
|
||||
<a class="sourceLine" id="cb2-26" title="26"><span class="co"># 6 10008 Male alive 62.8</span></a>
|
||||
<a class="sourceLine" id="cb2-27" title="27"><span class="co"># 7 10010 Female alive 63.7</span></a>
|
||||
<a class="sourceLine" id="cb2-28" title="28"><span class="co"># 8 10011 Male alive 73.1</span></a>
|
||||
<a class="sourceLine" id="cb2-29" title="29"><span class="co"># 9 10017 Male alive 56.7</span></a>
|
||||
<a class="sourceLine" id="cb2-30" title="30"><span class="co"># 10 10018 Female alive 66.6</span></a>
|
||||
<a class="sourceLine" id="cb2-31" title="31"><span class="co"># # … with 4,193 more rows</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb2"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb2-1" data-line-number="1">SPSS_data</a>
|
||||
<a class="sourceLine" id="cb2-2" data-line-number="2"><span class="co"># # A tibble: 4,203 x 4</span></a>
|
||||
<a class="sourceLine" id="cb2-3" data-line-number="3"><span class="co"># v001 sex status statusage</span></a>
|
||||
<a class="sourceLine" id="cb2-4" data-line-number="4"><span class="co"># <dbl> <dbl+lbl> <dbl+lbl> <dbl></span></a>
|
||||
<a class="sourceLine" id="cb2-5" data-line-number="5"><span class="co"># 1 10002 1 1 76.6</span></a>
|
||||
<a class="sourceLine" id="cb2-6" data-line-number="6"><span class="co"># 2 10004 0 1 59.1</span></a>
|
||||
<a class="sourceLine" id="cb2-7" data-line-number="7"><span class="co"># 3 10005 1 1 54.5</span></a>
|
||||
<a class="sourceLine" id="cb2-8" data-line-number="8"><span class="co"># 4 10006 1 1 54.1</span></a>
|
||||
<a class="sourceLine" id="cb2-9" data-line-number="9"><span class="co"># 5 10007 1 1 57.7</span></a>
|
||||
<a class="sourceLine" id="cb2-10" data-line-number="10"><span class="co"># 6 10008 1 1 62.8</span></a>
|
||||
<a class="sourceLine" id="cb2-11" data-line-number="11"><span class="co"># 7 10010 0 1 63.7</span></a>
|
||||
<a class="sourceLine" id="cb2-12" data-line-number="12"><span class="co"># 8 10011 1 1 73.1</span></a>
|
||||
<a class="sourceLine" id="cb2-13" data-line-number="13"><span class="co"># 9 10017 1 1 56.7</span></a>
|
||||
<a class="sourceLine" id="cb2-14" data-line-number="14"><span class="co"># 10 10018 0 1 66.6</span></a>
|
||||
<a class="sourceLine" id="cb2-15" data-line-number="15"><span class="co"># # … with 4,193 more rows</span></a>
|
||||
<a class="sourceLine" id="cb2-16" data-line-number="16"></a>
|
||||
<a class="sourceLine" id="cb2-17" data-line-number="17"><span class="kw">as_factor</span>(SPSS_data)</a>
|
||||
<a class="sourceLine" id="cb2-18" data-line-number="18"><span class="co"># # A tibble: 4,203 x 4</span></a>
|
||||
<a class="sourceLine" id="cb2-19" data-line-number="19"><span class="co"># v001 sex status statusage</span></a>
|
||||
<a class="sourceLine" id="cb2-20" data-line-number="20"><span class="co"># <dbl> <fct> <fct> <dbl></span></a>
|
||||
<a class="sourceLine" id="cb2-21" data-line-number="21"><span class="co"># 1 10002 Male alive 76.6</span></a>
|
||||
<a class="sourceLine" id="cb2-22" data-line-number="22"><span class="co"># 2 10004 Female alive 59.1</span></a>
|
||||
<a class="sourceLine" id="cb2-23" data-line-number="23"><span class="co"># 3 10005 Male alive 54.5</span></a>
|
||||
<a class="sourceLine" id="cb2-24" data-line-number="24"><span class="co"># 4 10006 Male alive 54.1</span></a>
|
||||
<a class="sourceLine" id="cb2-25" data-line-number="25"><span class="co"># 5 10007 Male alive 57.7</span></a>
|
||||
<a class="sourceLine" id="cb2-26" data-line-number="26"><span class="co"># 6 10008 Male alive 62.8</span></a>
|
||||
<a class="sourceLine" id="cb2-27" data-line-number="27"><span class="co"># 7 10010 Female alive 63.7</span></a>
|
||||
<a class="sourceLine" id="cb2-28" data-line-number="28"><span class="co"># 8 10011 Male alive 73.1</span></a>
|
||||
<a class="sourceLine" id="cb2-29" data-line-number="29"><span class="co"># 9 10017 Male alive 56.7</span></a>
|
||||
<a class="sourceLine" id="cb2-30" data-line-number="30"><span class="co"># 10 10018 Female alive 66.6</span></a>
|
||||
<a class="sourceLine" id="cb2-31" data-line-number="31"><span class="co"># # … with 4,193 more rows</span></a></code></pre></div>
|
||||
</div>
|
||||
<div id="base-r" class="section level3">
|
||||
<h3 class="hasAnchor">
|
||||
<a href="#base-r" class="anchor"></a>Base R</h3>
|
||||
<p>To import data from SPSS, SAS or Stata, you can use the <a href="https://haven.tidyverse.org/">great <code>haven</code> package</a> yourself:</p>
|
||||
<div class="sourceCode" id="cb3"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb3-1" title="1"><span class="co"># download and install the latest version:</span></a>
|
||||
<a class="sourceLine" id="cb3-2" title="2"><span class="kw"><a href="https://www.rdocumentation.org/packages/utils/topics/install.packages">install.packages</a></span>(<span class="st">"haven"</span>)</a>
|
||||
<a class="sourceLine" id="cb3-3" title="3"><span class="co"># load the package you just installed:</span></a>
|
||||
<a class="sourceLine" id="cb3-4" title="4"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(haven) </a></code></pre></div>
|
||||
<div class="sourceCode" id="cb3"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb3-1" data-line-number="1"><span class="co"># download and install the latest version:</span></a>
|
||||
<a class="sourceLine" id="cb3-2" data-line-number="2"><span class="kw"><a href="https://www.rdocumentation.org/packages/utils/topics/install.packages">install.packages</a></span>(<span class="st">"haven"</span>)</a>
|
||||
<a class="sourceLine" id="cb3-3" data-line-number="3"><span class="co"># load the package you just installed:</span></a>
|
||||
<a class="sourceLine" id="cb3-4" data-line-number="4"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(haven) </a></code></pre></div>
|
||||
<p>You can now import files as follows:</p>
|
||||
<div id="spss" class="section level4">
|
||||
<h4 class="hasAnchor">
|
||||
<a href="#spss" class="anchor"></a>SPSS</h4>
|
||||
<p>To read files from SPSS into R:</p>
|
||||
<div class="sourceCode" id="cb4"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb4-1" title="1"><span class="co"># read any SPSS file based on file extension (best way):</span></a>
|
||||
<a class="sourceLine" id="cb4-2" title="2"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_spss.html">read_spss</a></span>(<span class="dt">file =</span> <span class="st">"path/to/file"</span>)</a>
|
||||
<a class="sourceLine" id="cb4-3" title="3"></a>
|
||||
<a class="sourceLine" id="cb4-4" title="4"><span class="co"># read .sav or .zsav file:</span></a>
|
||||
<a class="sourceLine" id="cb4-5" title="5"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_spss.html">read_sav</a></span>(<span class="dt">file =</span> <span class="st">"path/to/file"</span>)</a>
|
||||
<a class="sourceLine" id="cb4-6" title="6"></a>
|
||||
<a class="sourceLine" id="cb4-7" title="7"><span class="co"># read .por file:</span></a>
|
||||
<a class="sourceLine" id="cb4-8" title="8"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_spss.html">read_por</a></span>(<span class="dt">file =</span> <span class="st">"path/to/file"</span>)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb4"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb4-1" data-line-number="1"><span class="co"># read any SPSS file based on file extension (best way):</span></a>
|
||||
<a class="sourceLine" id="cb4-2" data-line-number="2"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_spss.html">read_spss</a></span>(<span class="dt">file =</span> <span class="st">"path/to/file"</span>)</a>
|
||||
<a class="sourceLine" id="cb4-3" data-line-number="3"></a>
|
||||
<a class="sourceLine" id="cb4-4" data-line-number="4"><span class="co"># read .sav or .zsav file:</span></a>
|
||||
<a class="sourceLine" id="cb4-5" data-line-number="5"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_spss.html">read_sav</a></span>(<span class="dt">file =</span> <span class="st">"path/to/file"</span>)</a>
|
||||
<a class="sourceLine" id="cb4-6" data-line-number="6"></a>
|
||||
<a class="sourceLine" id="cb4-7" data-line-number="7"><span class="co"># read .por file:</span></a>
|
||||
<a class="sourceLine" id="cb4-8" data-line-number="8"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_spss.html">read_por</a></span>(<span class="dt">file =</span> <span class="st">"path/to/file"</span>)</a></code></pre></div>
|
||||
<p>Do not forget about <code><a href="https://haven.tidyverse.org/reference/as_factor.html">as_factor()</a></code>, as mentioned above.</p>
|
||||
<p>To export your R objects to the SPSS file format:</p>
|
||||
<div class="sourceCode" id="cb5"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb5-1" title="1"><span class="co"># save as .sav file:</span></a>
|
||||
<a class="sourceLine" id="cb5-2" title="2"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_spss.html">write_sav</a></span>(<span class="dt">data =</span> yourdata, <span class="dt">path =</span> <span class="st">"path/to/file"</span>)</a>
|
||||
<a class="sourceLine" id="cb5-3" title="3"></a>
|
||||
<a class="sourceLine" id="cb5-4" title="4"><span class="co"># save as compressed .zsav file:</span></a>
|
||||
<a class="sourceLine" id="cb5-5" title="5"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_spss.html">write_sav</a></span>(<span class="dt">data =</span> yourdata, <span class="dt">path =</span> <span class="st">"path/to/file"</span>, <span class="dt">compress =</span> <span class="ot">TRUE</span>)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb5"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb5-1" data-line-number="1"><span class="co"># save as .sav file:</span></a>
|
||||
<a class="sourceLine" id="cb5-2" data-line-number="2"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_spss.html">write_sav</a></span>(<span class="dt">data =</span> yourdata, <span class="dt">path =</span> <span class="st">"path/to/file"</span>)</a>
|
||||
<a class="sourceLine" id="cb5-3" data-line-number="3"></a>
|
||||
<a class="sourceLine" id="cb5-4" data-line-number="4"><span class="co"># save as compressed .zsav file:</span></a>
|
||||
<a class="sourceLine" id="cb5-5" data-line-number="5"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_spss.html">write_sav</a></span>(<span class="dt">data =</span> yourdata, <span class="dt">path =</span> <span class="st">"path/to/file"</span>, <span class="dt">compress =</span> <span class="ot">TRUE</span>)</a></code></pre></div>
|
||||
</div>
|
||||
<div id="sas" class="section level4">
|
||||
<h4 class="hasAnchor">
|
||||
<a href="#sas" class="anchor"></a>SAS</h4>
|
||||
<p>To read files from SAS into R:</p>
|
||||
<div class="sourceCode" id="cb6"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb6-1" title="1"><span class="co"># read .sas7bdat + .sas7bcat files:</span></a>
|
||||
<a class="sourceLine" id="cb6-2" title="2"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_sas.html">read_sas</a></span>(<span class="dt">data_file =</span> <span class="st">"path/to/file"</span>, <span class="dt">catalog_file =</span> <span class="ot">NULL</span>)</a>
|
||||
<a class="sourceLine" id="cb6-3" title="3"></a>
|
||||
<a class="sourceLine" id="cb6-4" title="4"><span class="co"># read SAS transport files (version 5 and version 8):</span></a>
|
||||
<a class="sourceLine" id="cb6-5" title="5"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_xpt.html">read_xpt</a></span>(<span class="dt">file =</span> <span class="st">"path/to/file"</span>)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb6"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb6-1" data-line-number="1"><span class="co"># read .sas7bdat + .sas7bcat files:</span></a>
|
||||
<a class="sourceLine" id="cb6-2" data-line-number="2"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_sas.html">read_sas</a></span>(<span class="dt">data_file =</span> <span class="st">"path/to/file"</span>, <span class="dt">catalog_file =</span> <span class="ot">NULL</span>)</a>
|
||||
<a class="sourceLine" id="cb6-3" data-line-number="3"></a>
|
||||
<a class="sourceLine" id="cb6-4" data-line-number="4"><span class="co"># read SAS transport files (version 5 and version 8):</span></a>
|
||||
<a class="sourceLine" id="cb6-5" data-line-number="5"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_xpt.html">read_xpt</a></span>(<span class="dt">file =</span> <span class="st">"path/to/file"</span>)</a></code></pre></div>
|
||||
<p>To export your R objects to the SAS file format:</p>
|
||||
<div class="sourceCode" id="cb7"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb7-1" title="1"><span class="co"># save as regular SAS file:</span></a>
|
||||
<a class="sourceLine" id="cb7-2" title="2"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_sas.html">write_sas</a></span>(<span class="dt">data =</span> yourdata, <span class="dt">path =</span> <span class="st">"path/to/file"</span>)</a>
|
||||
<a class="sourceLine" id="cb7-3" title="3"></a>
|
||||
<a class="sourceLine" id="cb7-4" title="4"><span class="co"># the SAS transport format is an open format </span></a>
|
||||
<a class="sourceLine" id="cb7-5" title="5"><span class="co"># (required for submission of the data to the FDA)</span></a>
|
||||
<a class="sourceLine" id="cb7-6" title="6"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_xpt.html">write_xpt</a></span>(<span class="dt">data =</span> yourdata, <span class="dt">path =</span> <span class="st">"path/to/file"</span>, <span class="dt">version =</span> <span class="dv">8</span>)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb7"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb7-1" data-line-number="1"><span class="co"># save as regular SAS file:</span></a>
|
||||
<a class="sourceLine" id="cb7-2" data-line-number="2"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_sas.html">write_sas</a></span>(<span class="dt">data =</span> yourdata, <span class="dt">path =</span> <span class="st">"path/to/file"</span>)</a>
|
||||
<a class="sourceLine" id="cb7-3" data-line-number="3"></a>
|
||||
<a class="sourceLine" id="cb7-4" data-line-number="4"><span class="co"># the SAS transport format is an open format </span></a>
|
||||
<a class="sourceLine" id="cb7-5" data-line-number="5"><span class="co"># (required for submission of the data to the FDA)</span></a>
|
||||
<a class="sourceLine" id="cb7-6" data-line-number="6"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_xpt.html">write_xpt</a></span>(<span class="dt">data =</span> yourdata, <span class="dt">path =</span> <span class="st">"path/to/file"</span>, <span class="dt">version =</span> <span class="dv">8</span>)</a></code></pre></div>
|
||||
</div>
|
||||
<div id="stata" class="section level4">
|
||||
<h4 class="hasAnchor">
|
||||
<a href="#stata" class="anchor"></a>Stata</h4>
|
||||
<p>To read files from Stata into R:</p>
|
||||
<div class="sourceCode" id="cb8"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb8-1" title="1"><span class="co"># read .dta file:</span></a>
|
||||
<a class="sourceLine" id="cb8-2" title="2"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_dta.html">read_stata</a></span>(<span class="dt">file =</span> <span class="st">"/path/to/file"</span>)</a>
|
||||
<a class="sourceLine" id="cb8-3" title="3"></a>
|
||||
<a class="sourceLine" id="cb8-4" title="4"><span class="co"># works exactly the same:</span></a>
|
||||
<a class="sourceLine" id="cb8-5" title="5"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_dta.html">read_dta</a></span>(<span class="dt">file =</span> <span class="st">"/path/to/file"</span>)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb8"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb8-1" data-line-number="1"><span class="co"># read .dta file:</span></a>
|
||||
<a class="sourceLine" id="cb8-2" data-line-number="2"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_dta.html">read_stata</a></span>(<span class="dt">file =</span> <span class="st">"/path/to/file"</span>)</a>
|
||||
<a class="sourceLine" id="cb8-3" data-line-number="3"></a>
|
||||
<a class="sourceLine" id="cb8-4" data-line-number="4"><span class="co"># works exactly the same:</span></a>
|
||||
<a class="sourceLine" id="cb8-5" data-line-number="5"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_dta.html">read_dta</a></span>(<span class="dt">file =</span> <span class="st">"/path/to/file"</span>)</a></code></pre></div>
|
||||
<p>To export your R objects to the Stata file format:</p>
|
||||
<div class="sourceCode" id="cb9"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb9-1" title="1"><span class="co"># save as .dta file, Stata version 14:</span></a>
|
||||
<a class="sourceLine" id="cb9-2" title="2"><span class="co"># (supports Stata v8 until v15 at the time of writing)</span></a>
|
||||
<a class="sourceLine" id="cb9-3" title="3"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_dta.html">write_dta</a></span>(<span class="dt">data =</span> yourdata, <span class="dt">path =</span> <span class="st">"/path/to/file"</span>, <span class="dt">version =</span> <span class="dv">14</span>)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb9"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb9-1" data-line-number="1"><span class="co"># save as .dta file, Stata version 14:</span></a>
|
||||
<a class="sourceLine" id="cb9-2" data-line-number="2"><span class="co"># (supports Stata v8 until v15 at the time of writing)</span></a>
|
||||
<a class="sourceLine" id="cb9-3" data-line-number="3"><span class="kw"><a href="https://haven.tidyverse.org/reference/read_dta.html">write_dta</a></span>(<span class="dt">data =</span> yourdata, <span class="dt">path =</span> <span class="st">"/path/to/file"</span>, <span class="dt">version =</span> <span class="dv">14</span>)</a></code></pre></div>
|
||||
</div>
|
||||
</div>
|
||||
</div>
|
||||
|
||||
@ -40,7 +40,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9026</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -118,13 +118,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -192,7 +185,7 @@
|
||||
<h1>How to work with WHONET data</h1>
|
||||
<h4 class="author">Matthijs S. Berends</h4>
|
||||
|
||||
<h4 class="date">01 July 2019</h4>
|
||||
<h4 class="date">06 August 2019</h4>
|
||||
|
||||
|
||||
<div class="hidden name"><code>WHONET.Rmd</code></div>
|
||||
@ -206,38 +199,42 @@
|
||||
<a href="#import-of-data" class="anchor"></a>Import of data</h1>
|
||||
<p>This tutorial assumes you already imported the WHONET data with e.g. the <a href="https://readxl.tidyverse.org/"><code>readxl</code> package</a>. In RStudio, this can be done using the menu button ‘Import Dataset’ in the tab ‘Environment’. Choose the option ‘From Excel’ and select your exported file. Make sure date fields are imported correctly.</p>
|
||||
<p>An example syntax could look like this:</p>
|
||||
<div class="sourceCode" id="cb1"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb1-1" title="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(readxl)</a>
|
||||
<a class="sourceLine" id="cb1-2" title="2">data <-<span class="st"> </span><span class="kw"><a href="https://readxl.tidyverse.org/reference/read_excel.html">read_excel</a></span>(<span class="dt">path =</span> <span class="st">"path/to/your/file.xlsx"</span>)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb1"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb1-1" data-line-number="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(readxl)</a>
|
||||
<a class="sourceLine" id="cb1-2" data-line-number="2">data <-<span class="st"> </span><span class="kw"><a href="https://readxl.tidyverse.org/reference/read_excel.html">read_excel</a></span>(<span class="dt">path =</span> <span class="st">"path/to/your/file.xlsx"</span>)</a></code></pre></div>
|
||||
<p>This package comes with an <a href="https://msberends.gitlab.io/AMR/reference/WHONET.html">example data set <code>WHONET</code></a>. We will use it for this analysis.</p>
|
||||
</div>
|
||||
<div id="preparation" class="section level1">
|
||||
<h1 class="hasAnchor">
|
||||
<a href="#preparation" class="anchor"></a>Preparation</h1>
|
||||
<p>First, load the relevant packages if you did not yet did this. I use the tidyverse for all of my analyses. All of them. If you don’t know it yet, I suggest you read about it on their website: <a href="https://www.tidyverse.org/" class="uri">https://www.tidyverse.org/</a>.</p>
|
||||
<div class="sourceCode" id="cb2"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb2-1" title="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(dplyr) <span class="co"># part of tidyverse</span></a>
|
||||
<a class="sourceLine" id="cb2-2" title="2"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(ggplot2) <span class="co"># part of tidyverse</span></a>
|
||||
<a class="sourceLine" id="cb2-3" title="3"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(AMR) <span class="co"># this package</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb2"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb2-1" data-line-number="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(dplyr) <span class="co"># part of tidyverse</span></a>
|
||||
<a class="sourceLine" id="cb2-2" data-line-number="2"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(ggplot2) <span class="co"># part of tidyverse</span></a>
|
||||
<a class="sourceLine" id="cb2-3" data-line-number="3"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(AMR) <span class="co"># this package</span></a></code></pre></div>
|
||||
<p>We will have to transform some variables to simplify and automate the analysis:</p>
|
||||
<ul>
|
||||
<li>Microorganisms should be transformed to our own microorganism IDs (called an <code>mo</code>) using <a href="https://msberends.gitlab.io/AMR/reference/ITIS.html">the ITIS reference data set</a>, which contains all ~20,000 microorganisms from the taxonomic kingdoms Bacteria, Fungi and Protozoa. We do the tranformation with <code><a href="../reference/as.mo.html">as.mo()</a></code>. This function also recognises almost all WHONET abbreviations of microorganisms.</li>
|
||||
<li>Antimicrobial results or interpretations have to be clean and valid. In other words, they should only contain values <code>"S"</code>, <code>"I"</code> or <code>"R"</code>. That is exactly where the <code><a href="../reference/as.rsi.html">as.rsi()</a></code> function is for.</li>
|
||||
</ul>
|
||||
<div class="sourceCode" id="cb3"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb3-1" title="1"><span class="co"># transform variables</span></a>
|
||||
<a class="sourceLine" id="cb3-2" title="2">data <-<span class="st"> </span>WHONET <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb3-3" title="3"><span class="st"> </span><span class="co"># get microbial ID based on given organism</span></a>
|
||||
<a class="sourceLine" id="cb3-4" title="4"><span class="st"> </span><span class="kw"><a href="https://dplyr.tidyverse.org/reference/mutate.html">mutate</a></span>(<span class="dt">mo =</span> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(Organism)) <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb3-5" title="5"><span class="st"> </span><span class="co"># transform everything from "AMP_ND10" to "CIP_EE" to the new `rsi` class</span></a>
|
||||
<a class="sourceLine" id="cb3-6" title="6"><span class="st"> </span><span class="kw"><a href="https://dplyr.tidyverse.org/reference/mutate_all.html">mutate_at</a></span>(<span class="kw"><a href="https://dplyr.tidyverse.org/reference/vars.html">vars</a></span>(AMP_ND10<span class="op">:</span>CIP_EE), as.rsi)</a></code></pre></div>
|
||||
<p>No errors or warnings, so all values are transformed succesfully. Let’s check it though, with a couple of frequency tables:</p>
|
||||
<div class="sourceCode" id="cb4"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb4-1" title="1"><span class="co"># our newly created `mo` variable</span></a>
|
||||
<a class="sourceLine" id="cb4-2" title="2">data <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(mo, <span class="dt">nmax =</span> <span class="dv">10</span>)</a></code></pre></div>
|
||||
<p><strong>Frequency table of <code>mo</code> from <code>data</code> (500 x 54)</strong></p>
|
||||
<div class="sourceCode" id="cb3"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb3-1" data-line-number="1"><span class="co"># transform variables</span></a>
|
||||
<a class="sourceLine" id="cb3-2" data-line-number="2">data <-<span class="st"> </span>WHONET <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb3-3" data-line-number="3"><span class="st"> </span><span class="co"># get microbial ID based on given organism</span></a>
|
||||
<a class="sourceLine" id="cb3-4" data-line-number="4"><span class="st"> </span><span class="kw"><a href="https://dplyr.tidyverse.org/reference/mutate.html">mutate</a></span>(<span class="dt">mo =</span> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(Organism)) <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb3-5" data-line-number="5"><span class="st"> </span><span class="co"># transform everything from "AMP_ND10" to "CIP_EE" to the new `rsi` class</span></a>
|
||||
<a class="sourceLine" id="cb3-6" data-line-number="6"><span class="st"> </span><span class="kw"><a href="https://dplyr.tidyverse.org/reference/mutate_all.html">mutate_at</a></span>(<span class="kw"><a href="https://dplyr.tidyverse.org/reference/vars.html">vars</a></span>(AMP_ND10<span class="op">:</span>CIP_EE), as.rsi)</a></code></pre></div>
|
||||
<p>No errors or warnings, so all values are transformed succesfully.</p>
|
||||
<p>We created a package dedicated to data cleaning and checking, called the <code>clean</code> package. It gets automatically installed with the <code>AMR</code> package, so we only have to load it:</p>
|
||||
<div class="sourceCode" id="cb4"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb4-1" data-line-number="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(clean)</a></code></pre></div>
|
||||
<p>It contains the <code><a href="https://www.rdocumentation.org/packages/clean/topics/freq">freq()</a></code> function, to create frequency tables.</p>
|
||||
<p>So let’s check our data, with a couple of frequency tables:</p>
|
||||
<div class="sourceCode" id="cb5"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb5-1" data-line-number="1"><span class="co"># our newly created `mo` variable</span></a>
|
||||
<a class="sourceLine" id="cb5-2" data-line-number="2">data <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/clean/topics/freq">freq</a></span>(mo, <span class="dt">nmax =</span> <span class="dv">10</span>)</a></code></pre></div>
|
||||
<p><strong>Frequency table</strong></p>
|
||||
<p>Class: mo (character)<br>
|
||||
Length: 500 (of which NA: 0 = 0.00%)<br>
|
||||
Unique: 39</p>
|
||||
<p>Families: 10<br>
|
||||
Genera: 17<br>
|
||||
Species: 38</p>
|
||||
Species: 39</p>
|
||||
<table class="table">
|
||||
<thead><tr class="header">
|
||||
<th align="left"></th>
|
||||
@ -331,18 +328,16 @@ Species: 38</p>
|
||||
</tbody>
|
||||
</table>
|
||||
<p>(omitted 29 entries, n = 57 [11.4%])</p>
|
||||
<div class="sourceCode" id="cb5"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb5-1" title="1"></a>
|
||||
<a class="sourceLine" id="cb5-2" title="2"><span class="co"># our transformed antibiotic columns</span></a>
|
||||
<a class="sourceLine" id="cb5-3" title="3"><span class="co"># amoxicillin/clavulanic acid (J01CR02) as an example</span></a>
|
||||
<a class="sourceLine" id="cb5-4" title="4">data <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(AMC_ND2)</a></code></pre></div>
|
||||
<p><strong>Frequency table of <code>AMC_ND2</code> from <code>data</code> (500 x 54)</strong></p>
|
||||
<div class="sourceCode" id="cb6"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb6-1" data-line-number="1"></a>
|
||||
<a class="sourceLine" id="cb6-2" data-line-number="2"><span class="co"># our transformed antibiotic columns</span></a>
|
||||
<a class="sourceLine" id="cb6-3" data-line-number="3"><span class="co"># amoxicillin/clavulanic acid (J01CR02) as an example</span></a>
|
||||
<a class="sourceLine" id="cb6-4" data-line-number="4">data <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/clean/topics/freq">freq</a></span>(AMC_ND2)</a></code></pre></div>
|
||||
<p><strong>Frequency table</strong></p>
|
||||
<p>Class: factor > ordered > rsi (numeric)<br>
|
||||
Length: 500 (of which NA: 19 = 3.80%)<br>
|
||||
Levels: 3: S < I < R<br>
|
||||
Unique: 3</p>
|
||||
<p>Drug: Amoxicillin/clavulanic acid (AMC, J01CR02)<br>
|
||||
Group: Beta-lactams/penicillins<br>
|
||||
%SI: 78.59%</p>
|
||||
<p>%SI: 78.6%</p>
|
||||
<table class="table">
|
||||
<thead><tr class="header">
|
||||
<th align="left"></th>
|
||||
|
||||
@ -40,7 +40,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -118,13 +118,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -192,7 +185,7 @@
|
||||
<h1>Benchmarks</h1>
|
||||
<h4 class="author">Matthijs S. Berends</h4>
|
||||
|
||||
<h4 class="date">01 July 2019</h4>
|
||||
<h4 class="date">29 July 2019</h4>
|
||||
|
||||
|
||||
<div class="hidden name"><code>benchmarks.Rmd</code></div>
|
||||
@ -203,161 +196,161 @@
|
||||
|
||||
<p>One of the most important features of this package is the complete microbial taxonomic database, supplied by the <a href="http://catalogueoflife.org">Catalogue of Life</a>. We created a function <code><a href="../reference/as.mo.html">as.mo()</a></code> that transforms any user input value to a valid microbial ID by using intelligent rules combined with the taxonomic tree of Catalogue of Life.</p>
|
||||
<p>Using the <code>microbenchmark</code> package, we can review the calculation performance of this function. Its function <code><a href="https://www.rdocumentation.org/packages/microbenchmark/topics/microbenchmark">microbenchmark()</a></code> runs different input expressions independently of each other and measures their time-to-result.</p>
|
||||
<div class="sourceCode" id="cb1"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb1-1" title="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(microbenchmark)</a>
|
||||
<a class="sourceLine" id="cb1-2" title="2"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(AMR)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb1"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb1-1" data-line-number="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(microbenchmark)</a>
|
||||
<a class="sourceLine" id="cb1-2" data-line-number="2"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(AMR)</a></code></pre></div>
|
||||
<p>In the next test, we try to ‘coerce’ different input values for <em>Staphylococcus aureus</em>. The actual result is the same every time: it returns its MO code <code>B_STPHY_AUR</code> (<em>B</em> stands for <em>Bacteria</em>, the taxonomic kingdom).</p>
|
||||
<p>But the calculation time differs a lot:</p>
|
||||
<div class="sourceCode" id="cb2"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb2-1" title="1">S.aureus <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/microbenchmark/topics/microbenchmark">microbenchmark</a></span>(<span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"sau"</span>),</a>
|
||||
<a class="sourceLine" id="cb2-2" title="2"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"stau"</span>),</a>
|
||||
<a class="sourceLine" id="cb2-3" title="3"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"staaur"</span>),</a>
|
||||
<a class="sourceLine" id="cb2-4" title="4"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"STAAUR"</span>),</a>
|
||||
<a class="sourceLine" id="cb2-5" title="5"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"S. aureus"</span>),</a>
|
||||
<a class="sourceLine" id="cb2-6" title="6"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"S. aureus"</span>),</a>
|
||||
<a class="sourceLine" id="cb2-7" title="7"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"Staphylococcus aureus"</span>),</a>
|
||||
<a class="sourceLine" id="cb2-8" title="8"> <span class="dt">times =</span> <span class="dv">10</span>)</a>
|
||||
<a class="sourceLine" id="cb2-9" title="9"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/print">print</a></span>(S.aureus, <span class="dt">unit =</span> <span class="st">"ms"</span>, <span class="dt">signif =</span> <span class="dv">2</span>)</a>
|
||||
<a class="sourceLine" id="cb2-10" title="10"><span class="co"># Unit: milliseconds</span></a>
|
||||
<a class="sourceLine" id="cb2-11" title="11"><span class="co"># expr min lq mean median uq max neval</span></a>
|
||||
<a class="sourceLine" id="cb2-12" title="12"><span class="co"># as.mo("sau") 18.0 18.0 22 18.0 18.0 61 10</span></a>
|
||||
<a class="sourceLine" id="cb2-13" title="13"><span class="co"># as.mo("stau") 65.0 65.0 70 66.0 66.0 110 10</span></a>
|
||||
<a class="sourceLine" id="cb2-14" title="14"><span class="co"># as.mo("staaur") 18.0 18.0 33 18.0 62.0 81 10</span></a>
|
||||
<a class="sourceLine" id="cb2-15" title="15"><span class="co"># as.mo("STAAUR") 18.0 18.0 18 18.0 18.0 19 10</span></a>
|
||||
<a class="sourceLine" id="cb2-16" title="16"><span class="co"># as.mo("S. aureus") 52.0 52.0 61 52.0 53.0 97 10</span></a>
|
||||
<a class="sourceLine" id="cb2-17" title="17"><span class="co"># as.mo("S. aureus") 52.0 52.0 71 53.0 97.0 150 10</span></a>
|
||||
<a class="sourceLine" id="cb2-18" title="18"><span class="co"># as.mo("Staphylococcus aureus") 8.1 8.1 14 8.1 8.2 63 10</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb2"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb2-1" data-line-number="1">S.aureus <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/microbenchmark/topics/microbenchmark">microbenchmark</a></span>(<span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"sau"</span>),</a>
|
||||
<a class="sourceLine" id="cb2-2" data-line-number="2"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"stau"</span>),</a>
|
||||
<a class="sourceLine" id="cb2-3" data-line-number="3"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"staaur"</span>),</a>
|
||||
<a class="sourceLine" id="cb2-4" data-line-number="4"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"STAAUR"</span>),</a>
|
||||
<a class="sourceLine" id="cb2-5" data-line-number="5"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"S. aureus"</span>),</a>
|
||||
<a class="sourceLine" id="cb2-6" data-line-number="6"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"S. aureus"</span>),</a>
|
||||
<a class="sourceLine" id="cb2-7" data-line-number="7"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"Staphylococcus aureus"</span>),</a>
|
||||
<a class="sourceLine" id="cb2-8" data-line-number="8"> <span class="dt">times =</span> <span class="dv">10</span>)</a>
|
||||
<a class="sourceLine" id="cb2-9" data-line-number="9"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/print">print</a></span>(S.aureus, <span class="dt">unit =</span> <span class="st">"ms"</span>, <span class="dt">signif =</span> <span class="dv">2</span>)</a>
|
||||
<a class="sourceLine" id="cb2-10" data-line-number="10"><span class="co"># Unit: milliseconds</span></a>
|
||||
<a class="sourceLine" id="cb2-11" data-line-number="11"><span class="co"># expr min lq mean median uq max neval</span></a>
|
||||
<a class="sourceLine" id="cb2-12" data-line-number="12"><span class="co"># as.mo("sau") 8.3 8.4 15.0 8.9 9.5 57.0 10</span></a>
|
||||
<a class="sourceLine" id="cb2-13" data-line-number="13"><span class="co"># as.mo("stau") 30.0 31.0 42.0 32.0 48.0 110.0 10</span></a>
|
||||
<a class="sourceLine" id="cb2-14" data-line-number="14"><span class="co"># as.mo("staaur") 8.3 8.4 10.0 8.5 8.9 25.0 10</span></a>
|
||||
<a class="sourceLine" id="cb2-15" data-line-number="15"><span class="co"># as.mo("STAAUR") 8.1 8.4 10.0 8.4 9.1 24.0 10</span></a>
|
||||
<a class="sourceLine" id="cb2-16" data-line-number="16"><span class="co"># as.mo("S. aureus") 23.0 23.0 29.0 24.0 38.0 45.0 10</span></a>
|
||||
<a class="sourceLine" id="cb2-17" data-line-number="17"><span class="co"># as.mo("S. aureus") 22.0 22.0 24.0 23.0 24.0 38.0 10</span></a>
|
||||
<a class="sourceLine" id="cb2-18" data-line-number="18"><span class="co"># as.mo("Staphylococcus aureus") 3.8 3.9 4.1 4.1 4.2 4.6 10</span></a></code></pre></div>
|
||||
<p>In the table above, all measurements are shown in milliseconds (thousands of seconds). A value of 5 milliseconds means it can determine 200 input values per second. It case of 100 milliseconds, this is only 10 input values per second. The second input is the only one that has to be looked up thoroughly. All the others are known codes (the first one is a WHONET code) or common laboratory codes, or common full organism names like the last one. Full organism names are always preferred.</p>
|
||||
<p>To achieve this speed, the <code>as.mo</code> function also takes into account the prevalence of human pathogenic microorganisms. The downside is of course that less prevalent microorganisms will be determined less fast. See this example for the ID of <em>Thermus islandicus</em> (<code>B_THERMS_ISL</code>), a bug probably never found before in humans:</p>
|
||||
<div class="sourceCode" id="cb3"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb3-1" title="1">T.islandicus <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/microbenchmark/topics/microbenchmark">microbenchmark</a></span>(<span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"theisl"</span>),</a>
|
||||
<a class="sourceLine" id="cb3-2" title="2"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"THEISL"</span>),</a>
|
||||
<a class="sourceLine" id="cb3-3" title="3"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"T. islandicus"</span>),</a>
|
||||
<a class="sourceLine" id="cb3-4" title="4"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"T. islandicus"</span>),</a>
|
||||
<a class="sourceLine" id="cb3-5" title="5"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"Thermus islandicus"</span>),</a>
|
||||
<a class="sourceLine" id="cb3-6" title="6"> <span class="dt">times =</span> <span class="dv">10</span>)</a>
|
||||
<a class="sourceLine" id="cb3-7" title="7"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/print">print</a></span>(T.islandicus, <span class="dt">unit =</span> <span class="st">"ms"</span>, <span class="dt">signif =</span> <span class="dv">2</span>)</a>
|
||||
<a class="sourceLine" id="cb3-8" title="8"><span class="co"># Unit: milliseconds</span></a>
|
||||
<a class="sourceLine" id="cb3-9" title="9"><span class="co"># expr min lq mean median uq max neval</span></a>
|
||||
<a class="sourceLine" id="cb3-10" title="10"><span class="co"># as.mo("theisl") 390 390 420 440 440 440 10</span></a>
|
||||
<a class="sourceLine" id="cb3-11" title="11"><span class="co"># as.mo("THEISL") 390 390 410 400 440 440 10</span></a>
|
||||
<a class="sourceLine" id="cb3-12" title="12"><span class="co"># as.mo("T. islandicus") 210 210 230 220 250 270 10</span></a>
|
||||
<a class="sourceLine" id="cb3-13" title="13"><span class="co"># as.mo("T. islandicus") 210 210 240 260 260 280 10</span></a>
|
||||
<a class="sourceLine" id="cb3-14" title="14"><span class="co"># as.mo("Thermus islandicus") 72 72 92 73 120 130 10</span></a></code></pre></div>
|
||||
<p>That takes 6.8 times as much time on average. A value of 100 milliseconds means it can only determine ~10 different input values per second. We can conclude that looking up arbitrary codes of less prevalent microorganisms is the worst way to go, in terms of calculation performance. Full names (like <em>Thermus islandicus</em>) are almost fast - these are the most probable input from most data sets.</p>
|
||||
<div class="sourceCode" id="cb3"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb3-1" data-line-number="1">T.islandicus <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/microbenchmark/topics/microbenchmark">microbenchmark</a></span>(<span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"theisl"</span>),</a>
|
||||
<a class="sourceLine" id="cb3-2" data-line-number="2"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"THEISL"</span>),</a>
|
||||
<a class="sourceLine" id="cb3-3" data-line-number="3"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"T. islandicus"</span>),</a>
|
||||
<a class="sourceLine" id="cb3-4" data-line-number="4"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"T. islandicus"</span>),</a>
|
||||
<a class="sourceLine" id="cb3-5" data-line-number="5"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"Thermus islandicus"</span>),</a>
|
||||
<a class="sourceLine" id="cb3-6" data-line-number="6"> <span class="dt">times =</span> <span class="dv">10</span>)</a>
|
||||
<a class="sourceLine" id="cb3-7" data-line-number="7"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/print">print</a></span>(T.islandicus, <span class="dt">unit =</span> <span class="st">"ms"</span>, <span class="dt">signif =</span> <span class="dv">2</span>)</a>
|
||||
<a class="sourceLine" id="cb3-8" data-line-number="8"><span class="co"># Unit: milliseconds</span></a>
|
||||
<a class="sourceLine" id="cb3-9" data-line-number="9"><span class="co"># expr min lq mean median uq max neval</span></a>
|
||||
<a class="sourceLine" id="cb3-10" data-line-number="10"><span class="co"># as.mo("theisl") 250 260 260 260 270 290 10</span></a>
|
||||
<a class="sourceLine" id="cb3-11" data-line-number="11"><span class="co"># as.mo("THEISL") 250 260 270 260 270 320 10</span></a>
|
||||
<a class="sourceLine" id="cb3-12" data-line-number="12"><span class="co"># as.mo("T. islandicus") 120 120 130 130 140 150 10</span></a>
|
||||
<a class="sourceLine" id="cb3-13" data-line-number="13"><span class="co"># as.mo("T. islandicus") 120 130 140 140 140 140 10</span></a>
|
||||
<a class="sourceLine" id="cb3-14" data-line-number="14"><span class="co"># as.mo("Thermus islandicus") 45 46 55 56 62 67 10</span></a></code></pre></div>
|
||||
<p>That takes 8.8 times as much time on average. A value of 100 milliseconds means it can only determine ~10 different input values per second. We can conclude that looking up arbitrary codes of less prevalent microorganisms is the worst way to go, in terms of calculation performance. Full names (like <em>Thermus islandicus</em>) are almost fast - these are the most probable input from most data sets.</p>
|
||||
<p>In the figure below, we compare <em>Escherichia coli</em> (which is very common) with <em>Prevotella brevis</em> (which is moderately common) and with <em>Thermus islandicus</em> (which is very uncommon):</p>
|
||||
<div class="sourceCode" id="cb4"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb4-1" title="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/graphics/topics/par">par</a></span>(<span class="dt">mar =</span> <span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/c">c</a></span>(<span class="dv">5</span>, <span class="dv">16</span>, <span class="dv">4</span>, <span class="dv">2</span>)) <span class="co"># set more space for left margin text (16)</span></a>
|
||||
<a class="sourceLine" id="cb4-2" title="2"></a>
|
||||
<a class="sourceLine" id="cb4-3" title="3"><span class="kw"><a href="https://www.rdocumentation.org/packages/graphics/topics/boxplot">boxplot</a></span>(<span class="kw"><a href="https://www.rdocumentation.org/packages/microbenchmark/topics/microbenchmark">microbenchmark</a></span>(<span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"Thermus islandicus"</span>),</a>
|
||||
<a class="sourceLine" id="cb4-4" title="4"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"Prevotella brevis"</span>),</a>
|
||||
<a class="sourceLine" id="cb4-5" title="5"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"Escherichia coli"</span>),</a>
|
||||
<a class="sourceLine" id="cb4-6" title="6"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"T. islandicus"</span>),</a>
|
||||
<a class="sourceLine" id="cb4-7" title="7"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"P. brevis"</span>),</a>
|
||||
<a class="sourceLine" id="cb4-8" title="8"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"E. coli"</span>),</a>
|
||||
<a class="sourceLine" id="cb4-9" title="9"> <span class="dt">times =</span> <span class="dv">10</span>),</a>
|
||||
<a class="sourceLine" id="cb4-10" title="10"> <span class="dt">horizontal =</span> <span class="ot">TRUE</span>, <span class="dt">las =</span> <span class="dv">1</span>, <span class="dt">unit =</span> <span class="st">"s"</span>, <span class="dt">log =</span> <span class="ot">FALSE</span>,</a>
|
||||
<a class="sourceLine" id="cb4-11" title="11"> <span class="dt">xlab =</span> <span class="st">""</span>, <span class="dt">ylab =</span> <span class="st">"Time in seconds"</span>,</a>
|
||||
<a class="sourceLine" id="cb4-12" title="12"> <span class="dt">main =</span> <span class="st">"Benchmarks per prevalence"</span>)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb4"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb4-1" data-line-number="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/graphics/topics/par">par</a></span>(<span class="dt">mar =</span> <span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/c">c</a></span>(<span class="dv">5</span>, <span class="dv">16</span>, <span class="dv">4</span>, <span class="dv">2</span>)) <span class="co"># set more space for left margin text (16)</span></a>
|
||||
<a class="sourceLine" id="cb4-2" data-line-number="2"></a>
|
||||
<a class="sourceLine" id="cb4-3" data-line-number="3"><span class="kw"><a href="https://www.rdocumentation.org/packages/graphics/topics/boxplot">boxplot</a></span>(<span class="kw"><a href="https://www.rdocumentation.org/packages/microbenchmark/topics/microbenchmark">microbenchmark</a></span>(<span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"Thermus islandicus"</span>),</a>
|
||||
<a class="sourceLine" id="cb4-4" data-line-number="4"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"Prevotella brevis"</span>),</a>
|
||||
<a class="sourceLine" id="cb4-5" data-line-number="5"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"Escherichia coli"</span>),</a>
|
||||
<a class="sourceLine" id="cb4-6" data-line-number="6"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"T. islandicus"</span>),</a>
|
||||
<a class="sourceLine" id="cb4-7" data-line-number="7"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"P. brevis"</span>),</a>
|
||||
<a class="sourceLine" id="cb4-8" data-line-number="8"> <span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"E. coli"</span>),</a>
|
||||
<a class="sourceLine" id="cb4-9" data-line-number="9"> <span class="dt">times =</span> <span class="dv">10</span>),</a>
|
||||
<a class="sourceLine" id="cb4-10" data-line-number="10"> <span class="dt">horizontal =</span> <span class="ot">TRUE</span>, <span class="dt">las =</span> <span class="dv">1</span>, <span class="dt">unit =</span> <span class="st">"s"</span>, <span class="dt">log =</span> <span class="ot">FALSE</span>,</a>
|
||||
<a class="sourceLine" id="cb4-11" data-line-number="11"> <span class="dt">xlab =</span> <span class="st">""</span>, <span class="dt">ylab =</span> <span class="st">"Time in seconds"</span>,</a>
|
||||
<a class="sourceLine" id="cb4-12" data-line-number="12"> <span class="dt">main =</span> <span class="st">"Benchmarks per prevalence"</span>)</a></code></pre></div>
|
||||
<p><img src="benchmarks_files/figure-html/unnamed-chunk-5-1.png" width="720"></p>
|
||||
<p>Uncommon microorganisms take a lot more time than common microorganisms. To relieve this pitfall and further improve performance, two important calculations take almost no time at all: <strong>repetitive results</strong> and <strong>already precalculated results</strong>.</p>
|
||||
<div id="repetitive-results" class="section level3">
|
||||
<h3 class="hasAnchor">
|
||||
<a href="#repetitive-results" class="anchor"></a>Repetitive results</h3>
|
||||
<p>Repetitive results are unique values that are present more than once. Unique values will only be calculated once by <code><a href="../reference/as.mo.html">as.mo()</a></code>. We will use <code><a href="../reference/mo_property.html">mo_fullname()</a></code> for this test - a helper function that returns the full microbial name (genus, species and possibly subspecies) which uses <code><a href="../reference/as.mo.html">as.mo()</a></code> internally.</p>
|
||||
<div class="sourceCode" id="cb5"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb5-1" title="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(dplyr)</a>
|
||||
<a class="sourceLine" id="cb5-2" title="2"><span class="co"># take all MO codes from the septic_patients data set</span></a>
|
||||
<a class="sourceLine" id="cb5-3" title="3">x <-<span class="st"> </span>septic_patients<span class="op">$</span>mo <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb5-4" title="4"><span class="st"> </span><span class="co"># keep only the unique ones</span></a>
|
||||
<a class="sourceLine" id="cb5-5" title="5"><span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/unique">unique</a></span>() <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb5-6" title="6"><span class="st"> </span><span class="co"># pick 50 of them at random</span></a>
|
||||
<a class="sourceLine" id="cb5-7" title="7"><span class="st"> </span><span class="kw"><a href="https://dplyr.tidyverse.org/reference/sample.html">sample</a></span>(<span class="dv">50</span>) <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb5-8" title="8"><span class="st"> </span><span class="co"># paste that 10,000 times</span></a>
|
||||
<a class="sourceLine" id="cb5-9" title="9"><span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/rep">rep</a></span>(<span class="dv">10000</span>) <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb5-10" title="10"><span class="st"> </span><span class="co"># scramble it</span></a>
|
||||
<a class="sourceLine" id="cb5-11" title="11"><span class="st"> </span><span class="kw"><a href="https://dplyr.tidyverse.org/reference/sample.html">sample</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-12" title="12"> </a>
|
||||
<a class="sourceLine" id="cb5-13" title="13"><span class="co"># got indeed 50 times 10,000 = half a million?</span></a>
|
||||
<a class="sourceLine" id="cb5-14" title="14"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/length">length</a></span>(x)</a>
|
||||
<a class="sourceLine" id="cb5-15" title="15"><span class="co"># [1] 500000</span></a>
|
||||
<a class="sourceLine" id="cb5-16" title="16"></a>
|
||||
<a class="sourceLine" id="cb5-17" title="17"><span class="co"># and how many unique values do we have?</span></a>
|
||||
<a class="sourceLine" id="cb5-18" title="18"><span class="kw"><a href="https://dplyr.tidyverse.org/reference/n_distinct.html">n_distinct</a></span>(x)</a>
|
||||
<a class="sourceLine" id="cb5-19" title="19"><span class="co"># [1] 50</span></a>
|
||||
<a class="sourceLine" id="cb5-20" title="20"></a>
|
||||
<a class="sourceLine" id="cb5-21" title="21"><span class="co"># now let's see:</span></a>
|
||||
<a class="sourceLine" id="cb5-22" title="22">run_it <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/microbenchmark/topics/microbenchmark">microbenchmark</a></span>(<span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(x),</a>
|
||||
<a class="sourceLine" id="cb5-23" title="23"> <span class="dt">times =</span> <span class="dv">10</span>)</a>
|
||||
<a class="sourceLine" id="cb5-24" title="24"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/print">print</a></span>(run_it, <span class="dt">unit =</span> <span class="st">"ms"</span>, <span class="dt">signif =</span> <span class="dv">3</span>)</a>
|
||||
<a class="sourceLine" id="cb5-25" title="25"><span class="co"># Unit: milliseconds</span></a>
|
||||
<a class="sourceLine" id="cb5-26" title="26"><span class="co"># expr min lq mean median uq max neval</span></a>
|
||||
<a class="sourceLine" id="cb5-27" title="27"><span class="co"># mo_fullname(x) 1050 1050 1100 1090 1120 1230 10</span></a></code></pre></div>
|
||||
<p>So transforming 500,000 values (!!) of 50 unique values only takes 1.09 seconds (1092 ms). You only lose time on your unique input values.</p>
|
||||
<div class="sourceCode" id="cb5"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb5-1" data-line-number="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(dplyr)</a>
|
||||
<a class="sourceLine" id="cb5-2" data-line-number="2"><span class="co"># take all MO codes from the septic_patients data set</span></a>
|
||||
<a class="sourceLine" id="cb5-3" data-line-number="3">x <-<span class="st"> </span>septic_patients<span class="op">$</span>mo <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb5-4" data-line-number="4"><span class="st"> </span><span class="co"># keep only the unique ones</span></a>
|
||||
<a class="sourceLine" id="cb5-5" data-line-number="5"><span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/unique">unique</a></span>() <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb5-6" data-line-number="6"><span class="st"> </span><span class="co"># pick 50 of them at random</span></a>
|
||||
<a class="sourceLine" id="cb5-7" data-line-number="7"><span class="st"> </span><span class="kw"><a href="https://dplyr.tidyverse.org/reference/sample.html">sample</a></span>(<span class="dv">50</span>) <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb5-8" data-line-number="8"><span class="st"> </span><span class="co"># paste that 10,000 times</span></a>
|
||||
<a class="sourceLine" id="cb5-9" data-line-number="9"><span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/rep">rep</a></span>(<span class="dv">10000</span>) <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb5-10" data-line-number="10"><span class="st"> </span><span class="co"># scramble it</span></a>
|
||||
<a class="sourceLine" id="cb5-11" data-line-number="11"><span class="st"> </span><span class="kw"><a href="https://dplyr.tidyverse.org/reference/sample.html">sample</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-12" data-line-number="12"> </a>
|
||||
<a class="sourceLine" id="cb5-13" data-line-number="13"><span class="co"># got indeed 50 times 10,000 = half a million?</span></a>
|
||||
<a class="sourceLine" id="cb5-14" data-line-number="14"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/length">length</a></span>(x)</a>
|
||||
<a class="sourceLine" id="cb5-15" data-line-number="15"><span class="co"># [1] 500000</span></a>
|
||||
<a class="sourceLine" id="cb5-16" data-line-number="16"></a>
|
||||
<a class="sourceLine" id="cb5-17" data-line-number="17"><span class="co"># and how many unique values do we have?</span></a>
|
||||
<a class="sourceLine" id="cb5-18" data-line-number="18"><span class="kw"><a href="https://dplyr.tidyverse.org/reference/n_distinct.html">n_distinct</a></span>(x)</a>
|
||||
<a class="sourceLine" id="cb5-19" data-line-number="19"><span class="co"># [1] 50</span></a>
|
||||
<a class="sourceLine" id="cb5-20" data-line-number="20"></a>
|
||||
<a class="sourceLine" id="cb5-21" data-line-number="21"><span class="co"># now let's see:</span></a>
|
||||
<a class="sourceLine" id="cb5-22" data-line-number="22">run_it <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/microbenchmark/topics/microbenchmark">microbenchmark</a></span>(<span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(x),</a>
|
||||
<a class="sourceLine" id="cb5-23" data-line-number="23"> <span class="dt">times =</span> <span class="dv">10</span>)</a>
|
||||
<a class="sourceLine" id="cb5-24" data-line-number="24"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/print">print</a></span>(run_it, <span class="dt">unit =</span> <span class="st">"ms"</span>, <span class="dt">signif =</span> <span class="dv">3</span>)</a>
|
||||
<a class="sourceLine" id="cb5-25" data-line-number="25"><span class="co"># Unit: milliseconds</span></a>
|
||||
<a class="sourceLine" id="cb5-26" data-line-number="26"><span class="co"># expr min lq mean median uq max neval</span></a>
|
||||
<a class="sourceLine" id="cb5-27" data-line-number="27"><span class="co"># mo_fullname(x) 586 611 623 619 638 671 10</span></a></code></pre></div>
|
||||
<p>So transforming 500,000 values (!!) of 50 unique values only takes 0.62 seconds (618 ms). You only lose time on your unique input values.</p>
|
||||
</div>
|
||||
<div id="precalculated-results" class="section level3">
|
||||
<h3 class="hasAnchor">
|
||||
<a href="#precalculated-results" class="anchor"></a>Precalculated results</h3>
|
||||
<p>What about precalculated results? If the input is an already precalculated result of a helper function like <code><a href="../reference/mo_property.html">mo_fullname()</a></code>, it almost doesn’t take any time at all (see ‘C’ below):</p>
|
||||
<div class="sourceCode" id="cb6"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb6-1" title="1">run_it <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/microbenchmark/topics/microbenchmark">microbenchmark</a></span>(<span class="dt">A =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"B_STPHY_AUR"</span>),</a>
|
||||
<a class="sourceLine" id="cb6-2" title="2"> <span class="dt">B =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"S. aureus"</span>),</a>
|
||||
<a class="sourceLine" id="cb6-3" title="3"> <span class="dt">C =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"Staphylococcus aureus"</span>),</a>
|
||||
<a class="sourceLine" id="cb6-4" title="4"> <span class="dt">times =</span> <span class="dv">10</span>)</a>
|
||||
<a class="sourceLine" id="cb6-5" title="5"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/print">print</a></span>(run_it, <span class="dt">unit =</span> <span class="st">"ms"</span>, <span class="dt">signif =</span> <span class="dv">3</span>)</a>
|
||||
<a class="sourceLine" id="cb6-6" title="6"><span class="co"># Unit: milliseconds</span></a>
|
||||
<a class="sourceLine" id="cb6-7" title="7"><span class="co"># expr min lq mean median uq max neval</span></a>
|
||||
<a class="sourceLine" id="cb6-8" title="8"><span class="co"># A 13.00 13.20 13.60 13.60 14.00 14.40 10</span></a>
|
||||
<a class="sourceLine" id="cb6-9" title="9"><span class="co"># B 49.40 50.00 57.50 51.90 52.40 103.00 10</span></a>
|
||||
<a class="sourceLine" id="cb6-10" title="10"><span class="co"># C 1.52 1.72 1.81 1.78 1.98 1.99 10</span></a></code></pre></div>
|
||||
<p>So going from <code><a href="../reference/mo_property.html">mo_fullname("Staphylococcus aureus")</a></code> to <code>"Staphylococcus aureus"</code> takes 0.0018 seconds - it doesn’t even start calculating <em>if the result would be the same as the expected resulting value</em>. That goes for all helper functions:</p>
|
||||
<div class="sourceCode" id="cb7"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb7-1" title="1">run_it <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/microbenchmark/topics/microbenchmark">microbenchmark</a></span>(<span class="dt">A =</span> <span class="kw"><a href="../reference/mo_property.html">mo_species</a></span>(<span class="st">"aureus"</span>),</a>
|
||||
<a class="sourceLine" id="cb7-2" title="2"> <span class="dt">B =</span> <span class="kw"><a href="../reference/mo_property.html">mo_genus</a></span>(<span class="st">"Staphylococcus"</span>),</a>
|
||||
<a class="sourceLine" id="cb7-3" title="3"> <span class="dt">C =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"Staphylococcus aureus"</span>),</a>
|
||||
<a class="sourceLine" id="cb7-4" title="4"> <span class="dt">D =</span> <span class="kw"><a href="../reference/mo_property.html">mo_family</a></span>(<span class="st">"Staphylococcaceae"</span>),</a>
|
||||
<a class="sourceLine" id="cb7-5" title="5"> <span class="dt">E =</span> <span class="kw"><a href="../reference/mo_property.html">mo_order</a></span>(<span class="st">"Bacillales"</span>),</a>
|
||||
<a class="sourceLine" id="cb7-6" title="6"> <span class="dt">F =</span> <span class="kw"><a href="../reference/mo_property.html">mo_class</a></span>(<span class="st">"Bacilli"</span>),</a>
|
||||
<a class="sourceLine" id="cb7-7" title="7"> <span class="dt">G =</span> <span class="kw"><a href="../reference/mo_property.html">mo_phylum</a></span>(<span class="st">"Firmicutes"</span>),</a>
|
||||
<a class="sourceLine" id="cb7-8" title="8"> <span class="dt">H =</span> <span class="kw"><a href="../reference/mo_property.html">mo_kingdom</a></span>(<span class="st">"Bacteria"</span>),</a>
|
||||
<a class="sourceLine" id="cb7-9" title="9"> <span class="dt">times =</span> <span class="dv">10</span>)</a>
|
||||
<a class="sourceLine" id="cb7-10" title="10"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/print">print</a></span>(run_it, <span class="dt">unit =</span> <span class="st">"ms"</span>, <span class="dt">signif =</span> <span class="dv">3</span>)</a>
|
||||
<a class="sourceLine" id="cb7-11" title="11"><span class="co"># Unit: milliseconds</span></a>
|
||||
<a class="sourceLine" id="cb7-12" title="12"><span class="co"># expr min lq mean median uq max neval</span></a>
|
||||
<a class="sourceLine" id="cb7-13" title="13"><span class="co"># A 0.612 0.623 0.685 0.653 0.789 0.814 10</span></a>
|
||||
<a class="sourceLine" id="cb7-14" title="14"><span class="co"># B 0.556 0.575 0.680 0.671 0.689 0.958 10</span></a>
|
||||
<a class="sourceLine" id="cb7-15" title="15"><span class="co"># C 1.520 1.710 1.800 1.820 1.950 1.970 10</span></a>
|
||||
<a class="sourceLine" id="cb7-16" title="16"><span class="co"># D 0.547 0.665 0.723 0.688 0.811 0.997 10</span></a>
|
||||
<a class="sourceLine" id="cb7-17" title="17"><span class="co"># E 0.490 0.541 0.633 0.629 0.748 0.756 10</span></a>
|
||||
<a class="sourceLine" id="cb7-18" title="18"><span class="co"># F 0.482 0.569 0.612 0.590 0.663 0.756 10</span></a>
|
||||
<a class="sourceLine" id="cb7-19" title="19"><span class="co"># G 0.551 0.558 0.601 0.586 0.632 0.735 10</span></a>
|
||||
<a class="sourceLine" id="cb7-20" title="20"><span class="co"># H 0.494 0.564 0.595 0.575 0.608 0.757 10</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb6"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb6-1" data-line-number="1">run_it <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/microbenchmark/topics/microbenchmark">microbenchmark</a></span>(<span class="dt">A =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"B_STPHY_AUR"</span>),</a>
|
||||
<a class="sourceLine" id="cb6-2" data-line-number="2"> <span class="dt">B =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"S. aureus"</span>),</a>
|
||||
<a class="sourceLine" id="cb6-3" data-line-number="3"> <span class="dt">C =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"Staphylococcus aureus"</span>),</a>
|
||||
<a class="sourceLine" id="cb6-4" data-line-number="4"> <span class="dt">times =</span> <span class="dv">10</span>)</a>
|
||||
<a class="sourceLine" id="cb6-5" data-line-number="5"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/print">print</a></span>(run_it, <span class="dt">unit =</span> <span class="st">"ms"</span>, <span class="dt">signif =</span> <span class="dv">3</span>)</a>
|
||||
<a class="sourceLine" id="cb6-6" data-line-number="6"><span class="co"># Unit: milliseconds</span></a>
|
||||
<a class="sourceLine" id="cb6-7" data-line-number="7"><span class="co"># expr min lq mean median uq max neval</span></a>
|
||||
<a class="sourceLine" id="cb6-8" data-line-number="8"><span class="co"># A 6.350 6.600 7.050 6.870 7.35 8.37 10</span></a>
|
||||
<a class="sourceLine" id="cb6-9" data-line-number="9"><span class="co"># B 21.300 21.500 25.300 22.200 22.70 48.20 10</span></a>
|
||||
<a class="sourceLine" id="cb6-10" data-line-number="10"><span class="co"># C 0.624 0.753 0.804 0.783 0.87 1.01 10</span></a></code></pre></div>
|
||||
<p>So going from <code><a href="../reference/mo_property.html">mo_fullname("Staphylococcus aureus")</a></code> to <code>"Staphylococcus aureus"</code> takes 0.0008 seconds - it doesn’t even start calculating <em>if the result would be the same as the expected resulting value</em>. That goes for all helper functions:</p>
|
||||
<div class="sourceCode" id="cb7"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb7-1" data-line-number="1">run_it <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/microbenchmark/topics/microbenchmark">microbenchmark</a></span>(<span class="dt">A =</span> <span class="kw"><a href="../reference/mo_property.html">mo_species</a></span>(<span class="st">"aureus"</span>),</a>
|
||||
<a class="sourceLine" id="cb7-2" data-line-number="2"> <span class="dt">B =</span> <span class="kw"><a href="../reference/mo_property.html">mo_genus</a></span>(<span class="st">"Staphylococcus"</span>),</a>
|
||||
<a class="sourceLine" id="cb7-3" data-line-number="3"> <span class="dt">C =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"Staphylococcus aureus"</span>),</a>
|
||||
<a class="sourceLine" id="cb7-4" data-line-number="4"> <span class="dt">D =</span> <span class="kw"><a href="../reference/mo_property.html">mo_family</a></span>(<span class="st">"Staphylococcaceae"</span>),</a>
|
||||
<a class="sourceLine" id="cb7-5" data-line-number="5"> <span class="dt">E =</span> <span class="kw"><a href="../reference/mo_property.html">mo_order</a></span>(<span class="st">"Bacillales"</span>),</a>
|
||||
<a class="sourceLine" id="cb7-6" data-line-number="6"> <span class="dt">F =</span> <span class="kw"><a href="../reference/mo_property.html">mo_class</a></span>(<span class="st">"Bacilli"</span>),</a>
|
||||
<a class="sourceLine" id="cb7-7" data-line-number="7"> <span class="dt">G =</span> <span class="kw"><a href="../reference/mo_property.html">mo_phylum</a></span>(<span class="st">"Firmicutes"</span>),</a>
|
||||
<a class="sourceLine" id="cb7-8" data-line-number="8"> <span class="dt">H =</span> <span class="kw"><a href="../reference/mo_property.html">mo_kingdom</a></span>(<span class="st">"Bacteria"</span>),</a>
|
||||
<a class="sourceLine" id="cb7-9" data-line-number="9"> <span class="dt">times =</span> <span class="dv">10</span>)</a>
|
||||
<a class="sourceLine" id="cb7-10" data-line-number="10"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/print">print</a></span>(run_it, <span class="dt">unit =</span> <span class="st">"ms"</span>, <span class="dt">signif =</span> <span class="dv">3</span>)</a>
|
||||
<a class="sourceLine" id="cb7-11" data-line-number="11"><span class="co"># Unit: milliseconds</span></a>
|
||||
<a class="sourceLine" id="cb7-12" data-line-number="12"><span class="co"># expr min lq mean median uq max neval</span></a>
|
||||
<a class="sourceLine" id="cb7-13" data-line-number="13"><span class="co"># A 0.436 0.454 0.460 0.460 0.462 0.491 10</span></a>
|
||||
<a class="sourceLine" id="cb7-14" data-line-number="14"><span class="co"># B 0.472 0.480 0.496 0.488 0.513 0.542 10</span></a>
|
||||
<a class="sourceLine" id="cb7-15" data-line-number="15"><span class="co"># C 0.657 0.672 0.757 0.750 0.797 0.952 10</span></a>
|
||||
<a class="sourceLine" id="cb7-16" data-line-number="16"><span class="co"># D 0.478 0.495 0.500 0.499 0.503 0.540 10</span></a>
|
||||
<a class="sourceLine" id="cb7-17" data-line-number="17"><span class="co"># E 0.436 0.446 0.456 0.448 0.455 0.507 10</span></a>
|
||||
<a class="sourceLine" id="cb7-18" data-line-number="18"><span class="co"># F 0.437 0.447 0.455 0.454 0.460 0.478 10</span></a>
|
||||
<a class="sourceLine" id="cb7-19" data-line-number="19"><span class="co"># G 0.428 0.441 0.449 0.447 0.455 0.477 10</span></a>
|
||||
<a class="sourceLine" id="cb7-20" data-line-number="20"><span class="co"># H 0.438 0.445 0.456 0.451 0.472 0.477 10</span></a></code></pre></div>
|
||||
<p>Of course, when running <code><a href="../reference/mo_property.html">mo_phylum("Firmicutes")</a></code> the function has zero knowledge about the actual microorganism, namely <em>S. aureus</em>. But since the result would be <code>"Firmicutes"</code> too, there is no point in calculating the result. And because this package ‘knows’ all phyla of all known bacteria (according to the Catalogue of Life), it can just return the initial value immediately.</p>
|
||||
</div>
|
||||
<div id="results-in-other-languages" class="section level3">
|
||||
<h3 class="hasAnchor">
|
||||
<a href="#results-in-other-languages" class="anchor"></a>Results in other languages</h3>
|
||||
<p>When the system language is non-English and supported by this <code>AMR</code> package, some functions will have a translated result. This almost does’t take extra time:</p>
|
||||
<div class="sourceCode" id="cb8"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb8-1" title="1"><span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"CoNS"</span>, <span class="dt">language =</span> <span class="st">"en"</span>) <span class="co"># or just mo_fullname("CoNS") on an English system</span></a>
|
||||
<a class="sourceLine" id="cb8-2" title="2"><span class="co"># [1] "Coagulase-negative Staphylococcus (CoNS)"</span></a>
|
||||
<a class="sourceLine" id="cb8-3" title="3"></a>
|
||||
<a class="sourceLine" id="cb8-4" title="4"><span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"CoNS"</span>, <span class="dt">language =</span> <span class="st">"es"</span>) <span class="co"># or just mo_fullname("CoNS") on a Spanish system</span></a>
|
||||
<a class="sourceLine" id="cb8-5" title="5"><span class="co"># [1] "Staphylococcus coagulasa negativo (SCN)"</span></a>
|
||||
<a class="sourceLine" id="cb8-6" title="6"></a>
|
||||
<a class="sourceLine" id="cb8-7" title="7"><span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"CoNS"</span>, <span class="dt">language =</span> <span class="st">"nl"</span>) <span class="co"># or just mo_fullname("CoNS") on a Dutch system</span></a>
|
||||
<a class="sourceLine" id="cb8-8" title="8"><span class="co"># [1] "Coagulase-negatieve Staphylococcus (CNS)"</span></a>
|
||||
<a class="sourceLine" id="cb8-9" title="9"></a>
|
||||
<a class="sourceLine" id="cb8-10" title="10">run_it <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/microbenchmark/topics/microbenchmark">microbenchmark</a></span>(<span class="dt">en =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"CoNS"</span>, <span class="dt">language =</span> <span class="st">"en"</span>),</a>
|
||||
<a class="sourceLine" id="cb8-11" title="11"> <span class="dt">de =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"CoNS"</span>, <span class="dt">language =</span> <span class="st">"de"</span>),</a>
|
||||
<a class="sourceLine" id="cb8-12" title="12"> <span class="dt">nl =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"CoNS"</span>, <span class="dt">language =</span> <span class="st">"nl"</span>),</a>
|
||||
<a class="sourceLine" id="cb8-13" title="13"> <span class="dt">es =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"CoNS"</span>, <span class="dt">language =</span> <span class="st">"es"</span>),</a>
|
||||
<a class="sourceLine" id="cb8-14" title="14"> <span class="dt">it =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"CoNS"</span>, <span class="dt">language =</span> <span class="st">"it"</span>),</a>
|
||||
<a class="sourceLine" id="cb8-15" title="15"> <span class="dt">fr =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"CoNS"</span>, <span class="dt">language =</span> <span class="st">"fr"</span>),</a>
|
||||
<a class="sourceLine" id="cb8-16" title="16"> <span class="dt">pt =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"CoNS"</span>, <span class="dt">language =</span> <span class="st">"pt"</span>),</a>
|
||||
<a class="sourceLine" id="cb8-17" title="17"> <span class="dt">times =</span> <span class="dv">10</span>)</a>
|
||||
<a class="sourceLine" id="cb8-18" title="18"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/print">print</a></span>(run_it, <span class="dt">unit =</span> <span class="st">"ms"</span>, <span class="dt">signif =</span> <span class="dv">4</span>)</a>
|
||||
<a class="sourceLine" id="cb8-19" title="19"><span class="co"># Unit: milliseconds</span></a>
|
||||
<a class="sourceLine" id="cb8-20" title="20"><span class="co"># expr min lq mean median uq max neval</span></a>
|
||||
<a class="sourceLine" id="cb8-21" title="21"><span class="co"># en 43.00 43.12 45.51 44.82 44.89 56.61 10</span></a>
|
||||
<a class="sourceLine" id="cb8-22" title="22"><span class="co"># de 46.47 46.99 52.11 47.57 48.11 93.77 10</span></a>
|
||||
<a class="sourceLine" id="cb8-23" title="23"><span class="co"># nl 60.86 62.72 67.57 63.69 63.99 108.20 10</span></a>
|
||||
<a class="sourceLine" id="cb8-24" title="24"><span class="co"># es 45.74 46.05 52.37 46.42 47.98 103.00 10</span></a>
|
||||
<a class="sourceLine" id="cb8-25" title="25"><span class="co"># it 45.84 45.89 51.90 47.66 47.73 94.83 10</span></a>
|
||||
<a class="sourceLine" id="cb8-26" title="26"><span class="co"># fr 45.97 46.92 47.44 47.76 47.86 48.49 10</span></a>
|
||||
<a class="sourceLine" id="cb8-27" title="27"><span class="co"># pt 45.93 46.77 47.36 47.77 47.93 48.12 10</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb8"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb8-1" data-line-number="1"><span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"CoNS"</span>, <span class="dt">language =</span> <span class="st">"en"</span>) <span class="co"># or just mo_fullname("CoNS") on an English system</span></a>
|
||||
<a class="sourceLine" id="cb8-2" data-line-number="2"><span class="co"># [1] "Coagulase-negative Staphylococcus (CoNS)"</span></a>
|
||||
<a class="sourceLine" id="cb8-3" data-line-number="3"></a>
|
||||
<a class="sourceLine" id="cb8-4" data-line-number="4"><span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"CoNS"</span>, <span class="dt">language =</span> <span class="st">"es"</span>) <span class="co"># or just mo_fullname("CoNS") on a Spanish system</span></a>
|
||||
<a class="sourceLine" id="cb8-5" data-line-number="5"><span class="co"># [1] "Staphylococcus coagulasa negativo (SCN)"</span></a>
|
||||
<a class="sourceLine" id="cb8-6" data-line-number="6"></a>
|
||||
<a class="sourceLine" id="cb8-7" data-line-number="7"><span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"CoNS"</span>, <span class="dt">language =</span> <span class="st">"nl"</span>) <span class="co"># or just mo_fullname("CoNS") on a Dutch system</span></a>
|
||||
<a class="sourceLine" id="cb8-8" data-line-number="8"><span class="co"># [1] "Coagulase-negatieve Staphylococcus (CNS)"</span></a>
|
||||
<a class="sourceLine" id="cb8-9" data-line-number="9"></a>
|
||||
<a class="sourceLine" id="cb8-10" data-line-number="10">run_it <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/microbenchmark/topics/microbenchmark">microbenchmark</a></span>(<span class="dt">en =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"CoNS"</span>, <span class="dt">language =</span> <span class="st">"en"</span>),</a>
|
||||
<a class="sourceLine" id="cb8-11" data-line-number="11"> <span class="dt">de =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"CoNS"</span>, <span class="dt">language =</span> <span class="st">"de"</span>),</a>
|
||||
<a class="sourceLine" id="cb8-12" data-line-number="12"> <span class="dt">nl =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"CoNS"</span>, <span class="dt">language =</span> <span class="st">"nl"</span>),</a>
|
||||
<a class="sourceLine" id="cb8-13" data-line-number="13"> <span class="dt">es =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"CoNS"</span>, <span class="dt">language =</span> <span class="st">"es"</span>),</a>
|
||||
<a class="sourceLine" id="cb8-14" data-line-number="14"> <span class="dt">it =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"CoNS"</span>, <span class="dt">language =</span> <span class="st">"it"</span>),</a>
|
||||
<a class="sourceLine" id="cb8-15" data-line-number="15"> <span class="dt">fr =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"CoNS"</span>, <span class="dt">language =</span> <span class="st">"fr"</span>),</a>
|
||||
<a class="sourceLine" id="cb8-16" data-line-number="16"> <span class="dt">pt =</span> <span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"CoNS"</span>, <span class="dt">language =</span> <span class="st">"pt"</span>),</a>
|
||||
<a class="sourceLine" id="cb8-17" data-line-number="17"> <span class="dt">times =</span> <span class="dv">10</span>)</a>
|
||||
<a class="sourceLine" id="cb8-18" data-line-number="18"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/print">print</a></span>(run_it, <span class="dt">unit =</span> <span class="st">"ms"</span>, <span class="dt">signif =</span> <span class="dv">4</span>)</a>
|
||||
<a class="sourceLine" id="cb8-19" data-line-number="19"><span class="co"># Unit: milliseconds</span></a>
|
||||
<a class="sourceLine" id="cb8-20" data-line-number="20"><span class="co"># expr min lq mean median uq max neval</span></a>
|
||||
<a class="sourceLine" id="cb8-21" data-line-number="21"><span class="co"># en 17.21 17.67 18.20 18.40 18.62 18.81 10</span></a>
|
||||
<a class="sourceLine" id="cb8-22" data-line-number="22"><span class="co"># de 18.76 19.01 21.59 19.45 19.82 41.96 10</span></a>
|
||||
<a class="sourceLine" id="cb8-23" data-line-number="23"><span class="co"># nl 24.15 24.57 29.11 25.67 26.49 45.25 10</span></a>
|
||||
<a class="sourceLine" id="cb8-24" data-line-number="24"><span class="co"># es 18.31 19.02 19.33 19.37 19.93 20.09 10</span></a>
|
||||
<a class="sourceLine" id="cb8-25" data-line-number="25"><span class="co"># it 18.96 19.27 23.48 19.58 20.91 41.27 10</span></a>
|
||||
<a class="sourceLine" id="cb8-26" data-line-number="26"><span class="co"># fr 18.33 18.80 19.46 19.27 19.97 21.10 10</span></a>
|
||||
<a class="sourceLine" id="cb8-27" data-line-number="27"><span class="co"># pt 18.89 19.50 20.54 19.70 20.36 27.83 10</span></a></code></pre></div>
|
||||
<p>Currently supported are German, Dutch, Spanish, Italian, French and Portuguese.</p>
|
||||
</div>
|
||||
</div>
|
||||
|
||||
|
Before 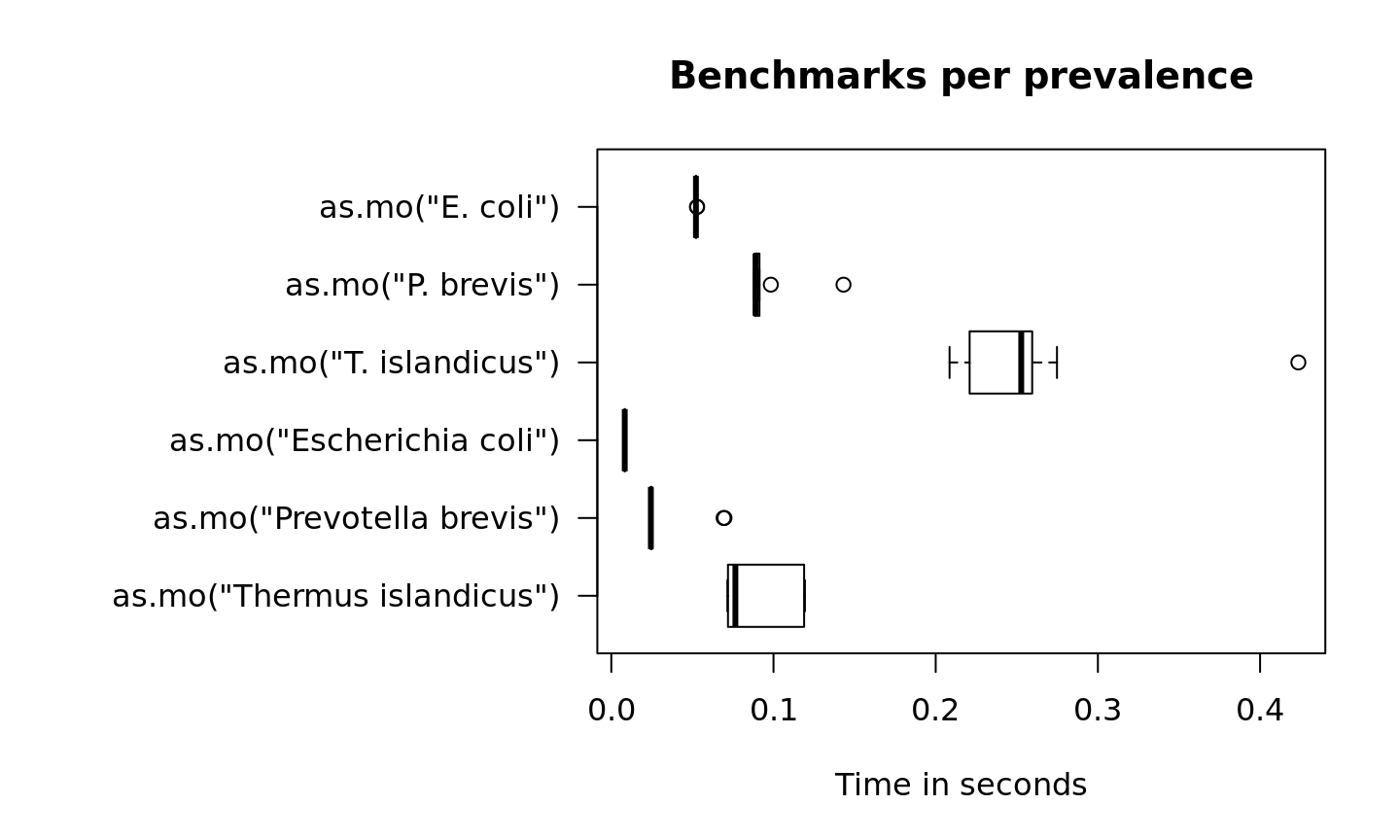
(image error) Size: 26 KiB After 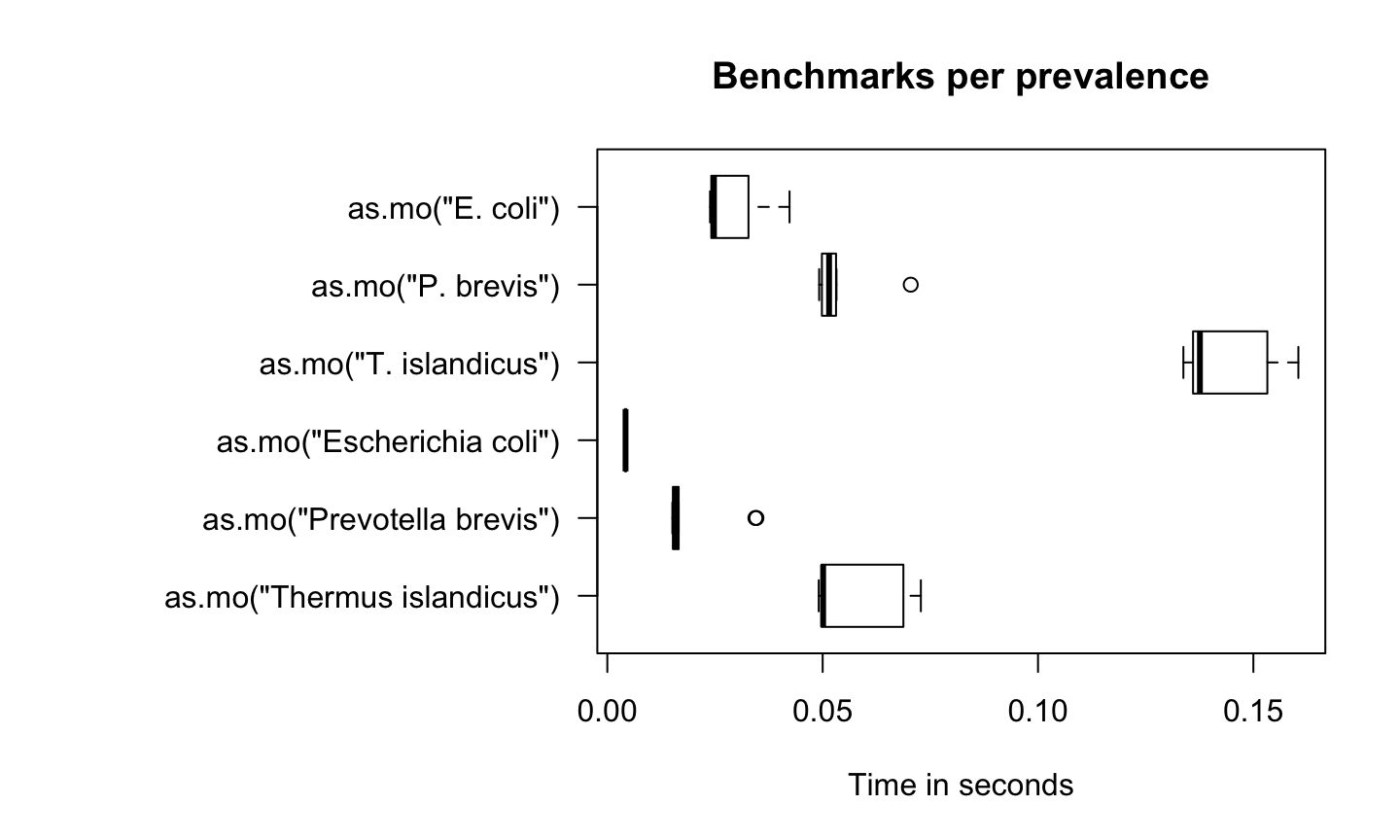
(image error) Size: 82 KiB 

|
@ -40,7 +40,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9012</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -192,7 +192,7 @@
|
||||
<h1>How to create frequency tables</h1>
|
||||
<h4 class="author">Matthijs S. Berends</h4>
|
||||
|
||||
<h4 class="date">01 July 2019</h4>
|
||||
<h4 class="date">10 July 2019</h4>
|
||||
|
||||
|
||||
<div class="hidden name"><code>freq.Rmd</code></div>
|
||||
@ -210,17 +210,17 @@
|
||||
<h2 class="hasAnchor">
|
||||
<a href="#frequencies-of-one-variable" class="anchor"></a>Frequencies of one variable</h2>
|
||||
<p>To only show and quickly review the content of one variable, you can just select this variable in various ways. Let’s say we want to get the frequencies of the <code>gender</code> variable of the <code>septic_patients</code> dataset:</p>
|
||||
<div class="sourceCode" id="cb1"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb1-1" title="1"><span class="co"># Any of these will work:</span></a>
|
||||
<a class="sourceLine" id="cb1-2" title="2"><span class="co"># freq(septic_patients$gender)</span></a>
|
||||
<a class="sourceLine" id="cb1-3" title="3"><span class="co"># freq(septic_patients[, "gender"])</span></a>
|
||||
<a class="sourceLine" id="cb1-4" title="4"></a>
|
||||
<a class="sourceLine" id="cb1-5" title="5"><span class="co"># Using tidyverse:</span></a>
|
||||
<a class="sourceLine" id="cb1-6" title="6"><span class="co"># septic_patients$gender %>% freq()</span></a>
|
||||
<a class="sourceLine" id="cb1-7" title="7"><span class="co"># septic_patients[, "gender"] %>% freq()</span></a>
|
||||
<a class="sourceLine" id="cb1-8" title="8"><span class="co"># septic_patients %>% freq("gender")</span></a>
|
||||
<a class="sourceLine" id="cb1-9" title="9"></a>
|
||||
<a class="sourceLine" id="cb1-10" title="10"><span class="co"># Probably the fastest and easiest:</span></a>
|
||||
<a class="sourceLine" id="cb1-11" title="11">septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(gender) </a></code></pre></div>
|
||||
<div class="sourceCode" id="cb1"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb1-1" data-line-number="1"><span class="co"># Any of these will work:</span></a>
|
||||
<a class="sourceLine" id="cb1-2" data-line-number="2"><span class="co"># freq(septic_patients$gender)</span></a>
|
||||
<a class="sourceLine" id="cb1-3" data-line-number="3"><span class="co"># freq(septic_patients[, "gender"])</span></a>
|
||||
<a class="sourceLine" id="cb1-4" data-line-number="4"></a>
|
||||
<a class="sourceLine" id="cb1-5" data-line-number="5"><span class="co"># Using tidyverse:</span></a>
|
||||
<a class="sourceLine" id="cb1-6" data-line-number="6"><span class="co"># septic_patients$gender %>% freq()</span></a>
|
||||
<a class="sourceLine" id="cb1-7" data-line-number="7"><span class="co"># septic_patients[, "gender"] %>% freq()</span></a>
|
||||
<a class="sourceLine" id="cb1-8" data-line-number="8"><span class="co"># septic_patients %>% freq("gender")</span></a>
|
||||
<a class="sourceLine" id="cb1-9" data-line-number="9"></a>
|
||||
<a class="sourceLine" id="cb1-10" data-line-number="10"><span class="co"># Probably the fastest and easiest:</span></a>
|
||||
<a class="sourceLine" id="cb1-11" data-line-number="11">septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(gender) </a></code></pre></div>
|
||||
<p><strong>Frequency table of <code>gender</code> from <code>septic_patients</code> (2,000 x 49)</strong></p>
|
||||
<p>Class: character<br>
|
||||
Length: 2,000 (of which NA: 0 = 0.00%)<br>
|
||||
@ -262,22 +262,22 @@ Longest: 1</p>
|
||||
<a href="#frequencies-of-more-than-one-variable" class="anchor"></a>Frequencies of more than one variable</h2>
|
||||
<p>Multiple variables will be pasted into one variable to review individual cases, keeping a univariate frequency table.</p>
|
||||
<p>For illustration, we could add some more variables to the <code>septic_patients</code> dataset to learn about bacterial properties:</p>
|
||||
<div class="sourceCode" id="cb2"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb2-1" title="1">my_patients <-<span class="st"> </span>septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/join.html">left_join_microorganisms</a></span>()</a>
|
||||
<a class="sourceLine" id="cb2-2" title="2"><span class="co"># Joining, by = "mo"</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb2"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb2-1" data-line-number="1">my_patients <-<span class="st"> </span>septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/join.html">left_join_microorganisms</a></span>()</a>
|
||||
<a class="sourceLine" id="cb2-2" data-line-number="2"><span class="co"># Joining, by = "mo"</span></a></code></pre></div>
|
||||
<p>Now all variables of the <code>microorganisms</code> dataset have been joined to the <code>septic_patients</code> dataset. The <code>microorganisms</code> dataset consists of the following variables:</p>
|
||||
<div class="sourceCode" id="cb3"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb3-1" title="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/colnames">colnames</a></span>(microorganisms)</a>
|
||||
<a class="sourceLine" id="cb3-2" title="2"><span class="co"># [1] "mo" "col_id" "fullname" "kingdom" "phylum" </span></a>
|
||||
<a class="sourceLine" id="cb3-3" title="3"><span class="co"># [6] "class" "order" "family" "genus" "species" </span></a>
|
||||
<a class="sourceLine" id="cb3-4" title="4"><span class="co"># [11] "subspecies" "rank" "ref" "species_id" "source" </span></a>
|
||||
<a class="sourceLine" id="cb3-5" title="5"><span class="co"># [16] "prevalence"</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb3"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb3-1" data-line-number="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/colnames">colnames</a></span>(microorganisms)</a>
|
||||
<a class="sourceLine" id="cb3-2" data-line-number="2"><span class="co"># [1] "mo" "col_id" "fullname" "kingdom" "phylum" </span></a>
|
||||
<a class="sourceLine" id="cb3-3" data-line-number="3"><span class="co"># [6] "class" "order" "family" "genus" "species" </span></a>
|
||||
<a class="sourceLine" id="cb3-4" data-line-number="4"><span class="co"># [11] "subspecies" "rank" "ref" "species_id" "source" </span></a>
|
||||
<a class="sourceLine" id="cb3-5" data-line-number="5"><span class="co"># [16] "prevalence"</span></a></code></pre></div>
|
||||
<p>If we compare the dimensions between the old and new dataset, we can see that these 15 variables were added:</p>
|
||||
<div class="sourceCode" id="cb4"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb4-1" title="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/dim">dim</a></span>(septic_patients)</a>
|
||||
<a class="sourceLine" id="cb4-2" title="2"><span class="co"># [1] 2000 49</span></a>
|
||||
<a class="sourceLine" id="cb4-3" title="3"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/dim">dim</a></span>(my_patients)</a>
|
||||
<a class="sourceLine" id="cb4-4" title="4"><span class="co"># [1] 2000 64</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb4"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb4-1" data-line-number="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/dim">dim</a></span>(septic_patients)</a>
|
||||
<a class="sourceLine" id="cb4-2" data-line-number="2"><span class="co"># [1] 2000 49</span></a>
|
||||
<a class="sourceLine" id="cb4-3" data-line-number="3"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/dim">dim</a></span>(my_patients)</a>
|
||||
<a class="sourceLine" id="cb4-4" data-line-number="4"><span class="co"># [1] 2000 64</span></a></code></pre></div>
|
||||
<p>So now the <code>genus</code> and <code>species</code> variables are available. A frequency table of these combined variables can be created like this:</p>
|
||||
<div class="sourceCode" id="cb5"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb5-1" title="1">my_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb5-2" title="2"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(genus, species, <span class="dt">nmax =</span> <span class="dv">15</span>)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb5"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb5-1" data-line-number="1">my_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb5-2" data-line-number="2"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(genus, species, <span class="dt">nmax =</span> <span class="dv">15</span>)</a></code></pre></div>
|
||||
<p><strong>Frequency table of <code>genus</code> and <code>species</code> from <code>my_patients</code> (2,000 x 64)</strong></p>
|
||||
<p>Columns: 2<br>
|
||||
Length: 2,000 (of which NA: 0 = 0.00%)<br>
|
||||
@ -423,10 +423,10 @@ Longest: 34</p>
|
||||
<a href="#frequencies-of-numeric-values" class="anchor"></a>Frequencies of numeric values</h2>
|
||||
<p>Frequency tables can be created of any input.</p>
|
||||
<p>In case of numeric values (like integers, doubles, etc.) additional descriptive statistics will be calculated and shown into the header:</p>
|
||||
<div class="sourceCode" id="cb6"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb6-1" title="1"><span class="co"># # get age distribution of unique patients</span></a>
|
||||
<a class="sourceLine" id="cb6-2" title="2">septic_patients <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb6-3" title="3"><span class="st"> </span><span class="kw">distinct</span>(patient_id, <span class="dt">.keep_all =</span> <span class="ot">TRUE</span>) <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb6-4" title="4"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(age, <span class="dt">nmax =</span> <span class="dv">5</span>, <span class="dt">header =</span> <span class="ot">TRUE</span>)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb6"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb6-1" data-line-number="1"><span class="co"># # get age distribution of unique patients</span></a>
|
||||
<a class="sourceLine" id="cb6-2" data-line-number="2">septic_patients <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb6-3" data-line-number="3"><span class="st"> </span><span class="kw">distinct</span>(patient_id, <span class="dt">.keep_all =</span> <span class="ot">TRUE</span>) <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb6-4" data-line-number="4"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(age, <span class="dt">nmax =</span> <span class="dv">5</span>, <span class="dt">header =</span> <span class="ot">TRUE</span>)</a></code></pre></div>
|
||||
<p><strong>Frequency table of <code>age</code> from a <code>data.frame</code> (981 x 49)</strong></p>
|
||||
<p>Class: numeric<br>
|
||||
Length: 981 (of which NA: 0 = 0.00%)<br>
|
||||
@ -506,8 +506,8 @@ Outliers: 15 (unique count: 12)</p>
|
||||
<a href="#frequencies-of-factors" class="anchor"></a>Frequencies of factors</h2>
|
||||
<p>To sort frequencies of factors on their levels instead of item count, use the <code>sort.count</code> parameter.</p>
|
||||
<p><code>sort.count</code> is <code>TRUE</code> by default. Compare this default behaviour…</p>
|
||||
<div class="sourceCode" id="cb7"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb7-1" title="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb7-2" title="2"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(hospital_id)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb7"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb7-1" data-line-number="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb7-2" data-line-number="2"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(hospital_id)</a></code></pre></div>
|
||||
<p><strong>Frequency table of <code>hospital_id</code> from <code>septic_patients</code> (2,000 x 49)</strong></p>
|
||||
<p>Class: factor (numeric)<br>
|
||||
Length: 2,000 (of which NA: 0 = 0.00%)<br>
|
||||
@ -558,8 +558,8 @@ Unique: 4</p>
|
||||
</tbody>
|
||||
</table>
|
||||
<p>… to this, where items are now sorted on factor levels:</p>
|
||||
<div class="sourceCode" id="cb8"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb8-1" title="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb8-2" title="2"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(hospital_id, <span class="dt">sort.count =</span> <span class="ot">FALSE</span>)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb8"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb8-1" data-line-number="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb8-2" data-line-number="2"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(hospital_id, <span class="dt">sort.count =</span> <span class="ot">FALSE</span>)</a></code></pre></div>
|
||||
<p><strong>Frequency table of <code>hospital_id</code> from <code>septic_patients</code> (2,000 x 49)</strong></p>
|
||||
<p>Class: factor (numeric)<br>
|
||||
Length: 2,000 (of which NA: 0 = 0.00%)<br>
|
||||
@ -610,8 +610,8 @@ Unique: 4</p>
|
||||
</tbody>
|
||||
</table>
|
||||
<p>All classes will be printed into the header. Variables with the new <code>rsi</code> class of this AMR package are actually ordered factors and have three classes (look at <code>Class</code> in the header):</p>
|
||||
<div class="sourceCode" id="cb9"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb9-1" title="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb9-2" title="2"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(AMX, <span class="dt">header =</span> <span class="ot">TRUE</span>)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb9"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb9-1" data-line-number="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb9-2" data-line-number="2"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(AMX, <span class="dt">header =</span> <span class="ot">TRUE</span>)</a></code></pre></div>
|
||||
<p><strong>Frequency table of <code>AMX</code> from <code>septic_patients</code> (2,000 x 49)</strong></p>
|
||||
<p>Class: factor > ordered > rsi (numeric)<br>
|
||||
Length: 2,000 (of which NA: 771 = 38.55%)<br>
|
||||
@ -661,8 +661,8 @@ Group: Beta-lactams/penicillins<br>
|
||||
<h2 class="hasAnchor">
|
||||
<a href="#frequencies-of-dates" class="anchor"></a>Frequencies of dates</h2>
|
||||
<p>Frequencies of dates will show the oldest and newest date in the data, and the amount of days between them:</p>
|
||||
<div class="sourceCode" id="cb10"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb10-1" title="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb10-2" title="2"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(date, <span class="dt">nmax =</span> <span class="dv">5</span>, <span class="dt">header =</span> <span class="ot">TRUE</span>)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb10"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb10-1" data-line-number="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb10-2" data-line-number="2"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(date, <span class="dt">nmax =</span> <span class="dv">5</span>, <span class="dt">header =</span> <span class="ot">TRUE</span>)</a></code></pre></div>
|
||||
<p><strong>Frequency table of <code>date</code> from <code>septic_patients</code> (2,000 x 49)</strong></p>
|
||||
<p>Class: Date (numeric)<br>
|
||||
Length: 2,000 (of which NA: 0 = 0.00%)<br>
|
||||
@ -728,11 +728,11 @@ Median: 31 July 2009 (47.39%)</p>
|
||||
<h2 class="hasAnchor">
|
||||
<a href="#assigning-a-frequency-table-to-an-object" class="anchor"></a>Assigning a frequency table to an object</h2>
|
||||
<p>A frequency table is actually a regular <code>data.frame</code>, with the exception that it contains an additional class.</p>
|
||||
<div class="sourceCode" id="cb11"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb11-1" title="1">my_df <-<span class="st"> </span>septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(age)</a>
|
||||
<a class="sourceLine" id="cb11-2" title="2"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/class">class</a></span>(my_df)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb11"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb11-1" data-line-number="1">my_df <-<span class="st"> </span>septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(age)</a>
|
||||
<a class="sourceLine" id="cb11-2" data-line-number="2"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/class">class</a></span>(my_df)</a></code></pre></div>
|
||||
<p>[1] “freq” “data.frame”</p>
|
||||
<p>Because of this additional class, a frequency table prints like the examples above. But the object itself contains the complete table without a row limitation:</p>
|
||||
<div class="sourceCode" id="cb12"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb12-1" title="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/dim">dim</a></span>(my_df)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb12"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb12-1" data-line-number="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/dim">dim</a></span>(my_df)</a></code></pre></div>
|
||||
<p>[1] 74 5</p>
|
||||
</div>
|
||||
<div id="additional-parameters" class="section level2">
|
||||
@ -743,8 +743,8 @@ Median: 31 July 2009 (47.39%)</p>
|
||||
<a href="#parameter-na-rm" class="anchor"></a>Parameter <code>na.rm</code>
|
||||
</h3>
|
||||
<p>With the <code>na.rm</code> parameter you can remove <code>NA</code> values from the frequency table (defaults to <code>TRUE</code>, but the number of <code>NA</code> values will always be shown into the header):</p>
|
||||
<div class="sourceCode" id="cb13"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb13-1" title="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb13-2" title="2"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(AMX, <span class="dt">na.rm =</span> <span class="ot">FALSE</span>)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb13"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb13-1" data-line-number="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb13-2" data-line-number="2"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(AMX, <span class="dt">na.rm =</span> <span class="ot">FALSE</span>)</a></code></pre></div>
|
||||
<p><strong>Frequency table of <code>AMX</code> from <code>septic_patients</code> (2,000 x 49)</strong></p>
|
||||
<p>Class: factor > ordered > rsi (numeric)<br>
|
||||
Length: 2,000 (of which NA: 771 = 38.55%)<br>
|
||||
@ -803,8 +803,8 @@ Group: Beta-lactams/penicillins<br>
|
||||
<a href="#parameter-row-names" class="anchor"></a>Parameter <code>row.names</code>
|
||||
</h3>
|
||||
<p>A frequency table shows row indices. To remove them, use <code>row.names = FALSE</code>:</p>
|
||||
<div class="sourceCode" id="cb14"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb14-1" title="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb14-2" title="2"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(hospital_id, <span class="dt">row.names =</span> <span class="ot">FALSE</span>)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb14"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb14-1" data-line-number="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb14-2" data-line-number="2"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(hospital_id, <span class="dt">row.names =</span> <span class="ot">FALSE</span>)</a></code></pre></div>
|
||||
<p><strong>Frequency table of <code>hospital_id</code> from <code>septic_patients</code> (2,000 x 49)</strong></p>
|
||||
<p>Class: factor (numeric)<br>
|
||||
Length: 2,000 (of which NA: 0 = 0.00%)<br>
|
||||
@ -855,21 +855,21 @@ Unique: 4</p>
|
||||
<a href="#parameter-markdown" class="anchor"></a>Parameter <code>markdown</code>
|
||||
</h3>
|
||||
<p>The <code>markdown</code> parameter is <code>TRUE</code> at default in non-interactive sessions, like in reports created with R Markdown. This will always print all rows, unless <code>nmax</code> is set. Without markdown (like in regular R), a frequency table would print like:</p>
|
||||
<div class="sourceCode" id="cb15"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb15-1" title="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb15-2" title="2"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(hospital_id, <span class="dt">markdown =</span> <span class="ot">FALSE</span>)</a>
|
||||
<a class="sourceLine" id="cb15-3" title="3"><span class="co"># Frequency table of `hospital_id` from `septic_patients` (2,000 x 49) </span></a>
|
||||
<a class="sourceLine" id="cb15-4" title="4"><span class="co"># </span></a>
|
||||
<a class="sourceLine" id="cb15-5" title="5"><span class="co"># Class: factor (numeric)</span></a>
|
||||
<a class="sourceLine" id="cb15-6" title="6"><span class="co"># Length: 2,000 (of which NA: 0 = 0.00%)</span></a>
|
||||
<a class="sourceLine" id="cb15-7" title="7"><span class="co"># Levels: 4: A, B, C, D</span></a>
|
||||
<a class="sourceLine" id="cb15-8" title="8"><span class="co"># Unique: 4</span></a>
|
||||
<a class="sourceLine" id="cb15-9" title="9"><span class="co"># </span></a>
|
||||
<a class="sourceLine" id="cb15-10" title="10"><span class="co"># Item Count Percent Cum. Count Cum. Percent</span></a>
|
||||
<a class="sourceLine" id="cb15-11" title="11"><span class="co"># --- ----- ------ -------- ----------- -------------</span></a>
|
||||
<a class="sourceLine" id="cb15-12" title="12"><span class="co"># 1 D 762 38.1% 762 38.1%</span></a>
|
||||
<a class="sourceLine" id="cb15-13" title="13"><span class="co"># 2 B 663 33.2% 1,425 71.2%</span></a>
|
||||
<a class="sourceLine" id="cb15-14" title="14"><span class="co"># 3 A 321 16.0% 1,746 87.3%</span></a>
|
||||
<a class="sourceLine" id="cb15-15" title="15"><span class="co"># 4 C 254 12.7% 2,000 100.0%</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb15"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb15-1" data-line-number="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb15-2" data-line-number="2"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(hospital_id, <span class="dt">markdown =</span> <span class="ot">FALSE</span>)</a>
|
||||
<a class="sourceLine" id="cb15-3" data-line-number="3"><span class="co"># Frequency table of `hospital_id` from `septic_patients` (2,000 x 49) </span></a>
|
||||
<a class="sourceLine" id="cb15-4" data-line-number="4"><span class="co"># </span></a>
|
||||
<a class="sourceLine" id="cb15-5" data-line-number="5"><span class="co"># Class: factor (numeric)</span></a>
|
||||
<a class="sourceLine" id="cb15-6" data-line-number="6"><span class="co"># Length: 2,000 (of which NA: 0 = 0.00%)</span></a>
|
||||
<a class="sourceLine" id="cb15-7" data-line-number="7"><span class="co"># Levels: 4: A, B, C, D</span></a>
|
||||
<a class="sourceLine" id="cb15-8" data-line-number="8"><span class="co"># Unique: 4</span></a>
|
||||
<a class="sourceLine" id="cb15-9" data-line-number="9"><span class="co"># </span></a>
|
||||
<a class="sourceLine" id="cb15-10" data-line-number="10"><span class="co"># Item Count Percent Cum. Count Cum. Percent</span></a>
|
||||
<a class="sourceLine" id="cb15-11" data-line-number="11"><span class="co"># --- ----- ------ -------- ----------- -------------</span></a>
|
||||
<a class="sourceLine" id="cb15-12" data-line-number="12"><span class="co"># 1 D 762 38.1% 762 38.1%</span></a>
|
||||
<a class="sourceLine" id="cb15-13" data-line-number="13"><span class="co"># 2 B 663 33.2% 1,425 71.2%</span></a>
|
||||
<a class="sourceLine" id="cb15-14" data-line-number="14"><span class="co"># 3 A 321 16.0% 1,746 87.3%</span></a>
|
||||
<a class="sourceLine" id="cb15-15" data-line-number="15"><span class="co"># 4 C 254 12.7% 2,000 100.0%</span></a></code></pre></div>
|
||||
</div>
|
||||
</div>
|
||||
</div>
|
||||
|
||||
@ -78,7 +78,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9008</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9026</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -156,13 +156,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -242,7 +235,6 @@
|
||||
<li><a href="SPSS.html">How to import data from SPSS / SAS / Stata</a></li>
|
||||
<li><a href="WHONET.html">How to work with WHONET data</a></li>
|
||||
<li><a href="benchmarks.html">Benchmarks</a></li>
|
||||
<li><a href="freq.html">How to create frequency tables</a></li>
|
||||
<li><a href="resistance_predict.html">How to predict antimicrobial resistance</a></li>
|
||||
</ul>
|
||||
</div>
|
||||
|
||||
@ -40,7 +40,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -118,13 +118,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -192,7 +185,7 @@
|
||||
<h1>How to predict antimicrobial resistance</h1>
|
||||
<h4 class="author">Matthijs S. Berends</h4>
|
||||
|
||||
<h4 class="date">01 July 2019</h4>
|
||||
<h4 class="date">29 July 2019</h4>
|
||||
|
||||
|
||||
<div class="hidden name"><code>resistance_predict.Rmd</code></div>
|
||||
@ -206,28 +199,28 @@
|
||||
<a href="#needed-r-packages" class="anchor"></a>Needed R packages</h2>
|
||||
<p>As with many uses in R, we need some additional packages for AMR analysis. Our package works closely together with the <a href="https://www.tidyverse.org">tidyverse packages</a> <a href="https://dplyr.tidyverse.org/"><code>dplyr</code></a> and <a href="https://ggplot2.tidyverse.org"><code>ggplot2</code></a> by Dr Hadley Wickham. The tidyverse tremendously improves the way we conduct data science - it allows for a very natural way of writing syntaxes and creating beautiful plots in R.</p>
|
||||
<p>Our <code>AMR</code> package depends on these packages and even extends their use and functions.</p>
|
||||
<div class="sourceCode" id="cb1"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb1-1" title="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(dplyr)</a>
|
||||
<a class="sourceLine" id="cb1-2" title="2"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(ggplot2)</a>
|
||||
<a class="sourceLine" id="cb1-3" title="3"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(AMR)</a>
|
||||
<a class="sourceLine" id="cb1-4" title="4"></a>
|
||||
<a class="sourceLine" id="cb1-5" title="5"><span class="co"># (if not yet installed, install with:)</span></a>
|
||||
<a class="sourceLine" id="cb1-6" title="6"><span class="co"># install.packages(c("tidyverse", "AMR"))</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb1"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb1-1" data-line-number="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(dplyr)</a>
|
||||
<a class="sourceLine" id="cb1-2" data-line-number="2"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(ggplot2)</a>
|
||||
<a class="sourceLine" id="cb1-3" data-line-number="3"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/library">library</a></span>(AMR)</a>
|
||||
<a class="sourceLine" id="cb1-4" data-line-number="4"></a>
|
||||
<a class="sourceLine" id="cb1-5" data-line-number="5"><span class="co"># (if not yet installed, install with:)</span></a>
|
||||
<a class="sourceLine" id="cb1-6" data-line-number="6"><span class="co"># install.packages(c("tidyverse", "AMR"))</span></a></code></pre></div>
|
||||
</div>
|
||||
<div id="prediction-analysis" class="section level2">
|
||||
<h2 class="hasAnchor">
|
||||
<a href="#prediction-analysis" class="anchor"></a>Prediction analysis</h2>
|
||||
<p>Our package contains a function <code><a href="../reference/resistance_predict.html">resistance_predict()</a></code>, which takes the same input as functions for <a href="./AMR.html">other AMR analysis</a>. Based on a date column, it calculates cases per year and uses a regression model to predict antimicrobial resistance.</p>
|
||||
<p>It is basically as easy as:</p>
|
||||
<div class="sourceCode" id="cb2"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb2-1" title="1"><span class="co"># resistance prediction of piperacillin/tazobactam (TZP):</span></a>
|
||||
<a class="sourceLine" id="cb2-2" title="2"><span class="kw"><a href="../reference/resistance_predict.html">resistance_predict</a></span>(<span class="dt">tbl =</span> septic_patients, <span class="dt">col_date =</span> <span class="st">"date"</span>, <span class="dt">col_ab =</span> <span class="st">"TZP"</span>)</a>
|
||||
<a class="sourceLine" id="cb2-3" title="3"></a>
|
||||
<a class="sourceLine" id="cb2-4" title="4"><span class="co"># or:</span></a>
|
||||
<a class="sourceLine" id="cb2-5" title="5">septic_patients <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb2-6" title="6"><span class="st"> </span><span class="kw"><a href="../reference/resistance_predict.html">resistance_predict</a></span>(<span class="dt">col_ab =</span> <span class="st">"TZP"</span>)</a>
|
||||
<a class="sourceLine" id="cb2-7" title="7"></a>
|
||||
<a class="sourceLine" id="cb2-8" title="8"><span class="co"># to bind it to object 'predict_TZP' for example:</span></a>
|
||||
<a class="sourceLine" id="cb2-9" title="9">predict_TZP <-<span class="st"> </span>septic_patients <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb2-10" title="10"><span class="st"> </span><span class="kw"><a href="../reference/resistance_predict.html">resistance_predict</a></span>(<span class="dt">col_ab =</span> <span class="st">"TZP"</span>)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb2"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb2-1" data-line-number="1"><span class="co"># resistance prediction of piperacillin/tazobactam (TZP):</span></a>
|
||||
<a class="sourceLine" id="cb2-2" data-line-number="2"><span class="kw"><a href="../reference/resistance_predict.html">resistance_predict</a></span>(<span class="dt">tbl =</span> septic_patients, <span class="dt">col_date =</span> <span class="st">"date"</span>, <span class="dt">col_ab =</span> <span class="st">"TZP"</span>)</a>
|
||||
<a class="sourceLine" id="cb2-3" data-line-number="3"></a>
|
||||
<a class="sourceLine" id="cb2-4" data-line-number="4"><span class="co"># or:</span></a>
|
||||
<a class="sourceLine" id="cb2-5" data-line-number="5">septic_patients <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb2-6" data-line-number="6"><span class="st"> </span><span class="kw"><a href="../reference/resistance_predict.html">resistance_predict</a></span>(<span class="dt">col_ab =</span> <span class="st">"TZP"</span>)</a>
|
||||
<a class="sourceLine" id="cb2-7" data-line-number="7"></a>
|
||||
<a class="sourceLine" id="cb2-8" data-line-number="8"><span class="co"># to bind it to object 'predict_TZP' for example:</span></a>
|
||||
<a class="sourceLine" id="cb2-9" data-line-number="9">predict_TZP <-<span class="st"> </span>septic_patients <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb2-10" data-line-number="10"><span class="st"> </span><span class="kw"><a href="../reference/resistance_predict.html">resistance_predict</a></span>(<span class="dt">col_ab =</span> <span class="st">"TZP"</span>)</a></code></pre></div>
|
||||
<p>The function will look for a date column itself if <code>col_date</code> is not set.</p>
|
||||
<p>When running any of these commands, a summary of the regression model will be printed unless using <code><a href="../reference/resistance_predict.html">resistance_predict(..., info = FALSE)</a></code>.</p>
|
||||
<pre><code># NOTE: Using column `date` as input for `col_date`.
|
||||
@ -257,55 +250,55 @@
|
||||
#
|
||||
# Number of Fisher Scoring iterations: 4</code></pre>
|
||||
<p>This text is only a printed summary - the actual result (output) of the function is a <code>data.frame</code> containing for each year: the number of observations, the actual observed resistance, the estimated resistance and the standard error below and above the estimation:</p>
|
||||
<div class="sourceCode" id="cb4"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb4-1" title="1">predict_TZP</a>
|
||||
<a class="sourceLine" id="cb4-2" title="2"><span class="co"># year value se_min se_max observations observed estimated</span></a>
|
||||
<a class="sourceLine" id="cb4-3" title="3"><span class="co"># 1 2003 0.06250000 NA NA 32 0.06250000 0.05486389</span></a>
|
||||
<a class="sourceLine" id="cb4-4" title="4"><span class="co"># 2 2004 0.08536585 NA NA 82 0.08536585 0.06089002</span></a>
|
||||
<a class="sourceLine" id="cb4-5" title="5"><span class="co"># 3 2005 0.05000000 NA NA 60 0.05000000 0.06753075</span></a>
|
||||
<a class="sourceLine" id="cb4-6" title="6"><span class="co"># 4 2006 0.05084746 NA NA 59 0.05084746 0.07483801</span></a>
|
||||
<a class="sourceLine" id="cb4-7" title="7"><span class="co"># 5 2007 0.12121212 NA NA 66 0.12121212 0.08286570</span></a>
|
||||
<a class="sourceLine" id="cb4-8" title="8"><span class="co"># 6 2008 0.04166667 NA NA 72 0.04166667 0.09166918</span></a>
|
||||
<a class="sourceLine" id="cb4-9" title="9"><span class="co"># 7 2009 0.01639344 NA NA 61 0.01639344 0.10130461</span></a>
|
||||
<a class="sourceLine" id="cb4-10" title="10"><span class="co"># 8 2010 0.05660377 NA NA 53 0.05660377 0.11182814</span></a>
|
||||
<a class="sourceLine" id="cb4-11" title="11"><span class="co"># 9 2011 0.18279570 NA NA 93 0.18279570 0.12329488</span></a>
|
||||
<a class="sourceLine" id="cb4-12" title="12"><span class="co"># 10 2012 0.30769231 NA NA 65 0.30769231 0.13575768</span></a>
|
||||
<a class="sourceLine" id="cb4-13" title="13"><span class="co"># 11 2013 0.06896552 NA NA 58 0.06896552 0.14926576</span></a>
|
||||
<a class="sourceLine" id="cb4-14" title="14"><span class="co"># 12 2014 0.10000000 NA NA 60 0.10000000 0.16386307</span></a>
|
||||
<a class="sourceLine" id="cb4-15" title="15"><span class="co"># 13 2015 0.23636364 NA NA 55 0.23636364 0.17958657</span></a>
|
||||
<a class="sourceLine" id="cb4-16" title="16"><span class="co"># 14 2016 0.22619048 NA NA 84 0.22619048 0.19646431</span></a>
|
||||
<a class="sourceLine" id="cb4-17" title="17"><span class="co"># 15 2017 0.16279070 NA NA 86 0.16279070 0.21451350</span></a>
|
||||
<a class="sourceLine" id="cb4-18" title="18"><span class="co"># 16 2018 0.23373852 0.2021578 0.2653193 NA NA 0.23373852</span></a>
|
||||
<a class="sourceLine" id="cb4-19" title="19"><span class="co"># 17 2019 0.25412909 0.2168525 0.2914057 NA NA 0.25412909</span></a>
|
||||
<a class="sourceLine" id="cb4-20" title="20"><span class="co"># 18 2020 0.27565854 0.2321869 0.3191302 NA NA 0.27565854</span></a>
|
||||
<a class="sourceLine" id="cb4-21" title="21"><span class="co"># 19 2021 0.29828252 0.2481942 0.3483709 NA NA 0.29828252</span></a>
|
||||
<a class="sourceLine" id="cb4-22" title="22"><span class="co"># 20 2022 0.32193804 0.2649008 0.3789753 NA NA 0.32193804</span></a>
|
||||
<a class="sourceLine" id="cb4-23" title="23"><span class="co"># 21 2023 0.34654311 0.2823269 0.4107593 NA NA 0.34654311</span></a>
|
||||
<a class="sourceLine" id="cb4-24" title="24"><span class="co"># 22 2024 0.37199700 0.3004860 0.4435080 NA NA 0.37199700</span></a>
|
||||
<a class="sourceLine" id="cb4-25" title="25"><span class="co"># 23 2025 0.39818127 0.3193839 0.4769787 NA NA 0.39818127</span></a>
|
||||
<a class="sourceLine" id="cb4-26" title="26"><span class="co"># 24 2026 0.42496142 0.3390173 0.5109056 NA NA 0.42496142</span></a>
|
||||
<a class="sourceLine" id="cb4-27" title="27"><span class="co"># 25 2027 0.45218939 0.3593720 0.5450068 NA NA 0.45218939</span></a>
|
||||
<a class="sourceLine" id="cb4-28" title="28"><span class="co"># 26 2028 0.47970658 0.3804212 0.5789920 NA NA 0.47970658</span></a>
|
||||
<a class="sourceLine" id="cb4-29" title="29"><span class="co"># 27 2029 0.50734745 0.4021241 0.6125708 NA NA 0.50734745</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb4"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb4-1" data-line-number="1">predict_TZP</a>
|
||||
<a class="sourceLine" id="cb4-2" data-line-number="2"><span class="co"># year value se_min se_max observations observed estimated</span></a>
|
||||
<a class="sourceLine" id="cb4-3" data-line-number="3"><span class="co"># 1 2003 0.06250000 NA NA 32 0.06250000 0.05486389</span></a>
|
||||
<a class="sourceLine" id="cb4-4" data-line-number="4"><span class="co"># 2 2004 0.08536585 NA NA 82 0.08536585 0.06089002</span></a>
|
||||
<a class="sourceLine" id="cb4-5" data-line-number="5"><span class="co"># 3 2005 0.05000000 NA NA 60 0.05000000 0.06753075</span></a>
|
||||
<a class="sourceLine" id="cb4-6" data-line-number="6"><span class="co"># 4 2006 0.05084746 NA NA 59 0.05084746 0.07483801</span></a>
|
||||
<a class="sourceLine" id="cb4-7" data-line-number="7"><span class="co"># 5 2007 0.12121212 NA NA 66 0.12121212 0.08286570</span></a>
|
||||
<a class="sourceLine" id="cb4-8" data-line-number="8"><span class="co"># 6 2008 0.04166667 NA NA 72 0.04166667 0.09166918</span></a>
|
||||
<a class="sourceLine" id="cb4-9" data-line-number="9"><span class="co"># 7 2009 0.01639344 NA NA 61 0.01639344 0.10130461</span></a>
|
||||
<a class="sourceLine" id="cb4-10" data-line-number="10"><span class="co"># 8 2010 0.05660377 NA NA 53 0.05660377 0.11182814</span></a>
|
||||
<a class="sourceLine" id="cb4-11" data-line-number="11"><span class="co"># 9 2011 0.18279570 NA NA 93 0.18279570 0.12329488</span></a>
|
||||
<a class="sourceLine" id="cb4-12" data-line-number="12"><span class="co"># 10 2012 0.30769231 NA NA 65 0.30769231 0.13575768</span></a>
|
||||
<a class="sourceLine" id="cb4-13" data-line-number="13"><span class="co"># 11 2013 0.06896552 NA NA 58 0.06896552 0.14926576</span></a>
|
||||
<a class="sourceLine" id="cb4-14" data-line-number="14"><span class="co"># 12 2014 0.10000000 NA NA 60 0.10000000 0.16386307</span></a>
|
||||
<a class="sourceLine" id="cb4-15" data-line-number="15"><span class="co"># 13 2015 0.23636364 NA NA 55 0.23636364 0.17958657</span></a>
|
||||
<a class="sourceLine" id="cb4-16" data-line-number="16"><span class="co"># 14 2016 0.22619048 NA NA 84 0.22619048 0.19646431</span></a>
|
||||
<a class="sourceLine" id="cb4-17" data-line-number="17"><span class="co"># 15 2017 0.16279070 NA NA 86 0.16279070 0.21451350</span></a>
|
||||
<a class="sourceLine" id="cb4-18" data-line-number="18"><span class="co"># 16 2018 0.23373852 0.2021578 0.2653193 NA NA 0.23373852</span></a>
|
||||
<a class="sourceLine" id="cb4-19" data-line-number="19"><span class="co"># 17 2019 0.25412909 0.2168525 0.2914057 NA NA 0.25412909</span></a>
|
||||
<a class="sourceLine" id="cb4-20" data-line-number="20"><span class="co"># 18 2020 0.27565854 0.2321869 0.3191302 NA NA 0.27565854</span></a>
|
||||
<a class="sourceLine" id="cb4-21" data-line-number="21"><span class="co"># 19 2021 0.29828252 0.2481942 0.3483709 NA NA 0.29828252</span></a>
|
||||
<a class="sourceLine" id="cb4-22" data-line-number="22"><span class="co"># 20 2022 0.32193804 0.2649008 0.3789753 NA NA 0.32193804</span></a>
|
||||
<a class="sourceLine" id="cb4-23" data-line-number="23"><span class="co"># 21 2023 0.34654311 0.2823269 0.4107593 NA NA 0.34654311</span></a>
|
||||
<a class="sourceLine" id="cb4-24" data-line-number="24"><span class="co"># 22 2024 0.37199700 0.3004860 0.4435080 NA NA 0.37199700</span></a>
|
||||
<a class="sourceLine" id="cb4-25" data-line-number="25"><span class="co"># 23 2025 0.39818127 0.3193839 0.4769787 NA NA 0.39818127</span></a>
|
||||
<a class="sourceLine" id="cb4-26" data-line-number="26"><span class="co"># 24 2026 0.42496142 0.3390173 0.5109056 NA NA 0.42496142</span></a>
|
||||
<a class="sourceLine" id="cb4-27" data-line-number="27"><span class="co"># 25 2027 0.45218939 0.3593720 0.5450068 NA NA 0.45218939</span></a>
|
||||
<a class="sourceLine" id="cb4-28" data-line-number="28"><span class="co"># 26 2028 0.47970658 0.3804212 0.5789920 NA NA 0.47970658</span></a>
|
||||
<a class="sourceLine" id="cb4-29" data-line-number="29"><span class="co"># 27 2029 0.50734745 0.4021241 0.6125708 NA NA 0.50734745</span></a></code></pre></div>
|
||||
<p>The function <code>plot</code> is available in base R, and can be extended by other packages to depend the output based on the type of input. We extended its function to cope with resistance predictions:</p>
|
||||
<div class="sourceCode" id="cb5"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb5-1" title="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/graphics/topics/plot">plot</a></span>(predict_TZP)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb5"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb5-1" data-line-number="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/graphics/topics/plot">plot</a></span>(predict_TZP)</a></code></pre></div>
|
||||
<p><img src="resistance_predict_files/figure-html/unnamed-chunk-4-1.png" width="720"></p>
|
||||
<p>This is the fastest way to plot the result. It automatically adds the right axes, error bars, titles, number of available observations and type of model.</p>
|
||||
<p>We also support the <code>ggplot2</code> package with our custom function <code><a href="../reference/resistance_predict.html">ggplot_rsi_predict()</a></code> to create more appealing plots:</p>
|
||||
<div class="sourceCode" id="cb6"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb6-1" title="1"><span class="kw"><a href="../reference/resistance_predict.html">ggplot_rsi_predict</a></span>(predict_TZP)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb6"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb6-1" data-line-number="1"><span class="kw"><a href="../reference/resistance_predict.html">ggplot_rsi_predict</a></span>(predict_TZP)</a></code></pre></div>
|
||||
<p><img src="resistance_predict_files/figure-html/unnamed-chunk-5-1.png" width="720"></p>
|
||||
<div class="sourceCode" id="cb7"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb7-1" title="1"></a>
|
||||
<a class="sourceLine" id="cb7-2" title="2"><span class="co"># choose for error bars instead of a ribbon</span></a>
|
||||
<a class="sourceLine" id="cb7-3" title="3"><span class="kw"><a href="../reference/resistance_predict.html">ggplot_rsi_predict</a></span>(predict_TZP, <span class="dt">ribbon =</span> <span class="ot">FALSE</span>)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb7"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb7-1" data-line-number="1"></a>
|
||||
<a class="sourceLine" id="cb7-2" data-line-number="2"><span class="co"># choose for error bars instead of a ribbon</span></a>
|
||||
<a class="sourceLine" id="cb7-3" data-line-number="3"><span class="kw"><a href="../reference/resistance_predict.html">ggplot_rsi_predict</a></span>(predict_TZP, <span class="dt">ribbon =</span> <span class="ot">FALSE</span>)</a></code></pre></div>
|
||||
<p><img src="resistance_predict_files/figure-html/unnamed-chunk-5-2.png" width="720"></p>
|
||||
<div id="choosing-the-right-model" class="section level3">
|
||||
<h3 class="hasAnchor">
|
||||
<a href="#choosing-the-right-model" class="anchor"></a>Choosing the right model</h3>
|
||||
<p>Resistance is not easily predicted; if we look at vancomycin resistance in Gram positives, the spread (i.e. standard error) is enormous:</p>
|
||||
<div class="sourceCode" id="cb8"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb8-1" title="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb8-2" title="2"><span class="st"> </span><span class="kw"><a href="https://dplyr.tidyverse.org/reference/filter.html">filter</a></span>(<span class="kw"><a href="../reference/mo_property.html">mo_gramstain</a></span>(mo, <span class="dt">language =</span> <span class="ot">NULL</span>) <span class="op">==</span><span class="st"> "Gram-positive"</span>) <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb8-3" title="3"><span class="st"> </span><span class="kw"><a href="../reference/resistance_predict.html">resistance_predict</a></span>(<span class="dt">col_ab =</span> <span class="st">"VAN"</span>, <span class="dt">year_min =</span> <span class="dv">2010</span>, <span class="dt">info =</span> <span class="ot">FALSE</span>) <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb8-4" title="4"><span class="st"> </span><span class="kw"><a href="../reference/resistance_predict.html">ggplot_rsi_predict</a></span>()</a>
|
||||
<a class="sourceLine" id="cb8-5" title="5"><span class="co"># </span><span class="al">NOTE</span><span class="co">: Using column `date` as input for `col_date`.</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb8"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb8-1" data-line-number="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb8-2" data-line-number="2"><span class="st"> </span><span class="kw"><a href="https://dplyr.tidyverse.org/reference/filter.html">filter</a></span>(<span class="kw"><a href="../reference/mo_property.html">mo_gramstain</a></span>(mo, <span class="dt">language =</span> <span class="ot">NULL</span>) <span class="op">==</span><span class="st"> "Gram-positive"</span>) <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb8-3" data-line-number="3"><span class="st"> </span><span class="kw"><a href="../reference/resistance_predict.html">resistance_predict</a></span>(<span class="dt">col_ab =</span> <span class="st">"VAN"</span>, <span class="dt">year_min =</span> <span class="dv">2010</span>, <span class="dt">info =</span> <span class="ot">FALSE</span>) <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb8-4" data-line-number="4"><span class="st"> </span><span class="kw"><a href="../reference/resistance_predict.html">ggplot_rsi_predict</a></span>()</a>
|
||||
<a class="sourceLine" id="cb8-5" data-line-number="5"><span class="co"># </span><span class="al">NOTE</span><span class="co">: Using column `date` as input for `col_date`.</span></a></code></pre></div>
|
||||
<p><img src="resistance_predict_files/figure-html/unnamed-chunk-6-1.png" width="720"></p>
|
||||
<p>Vancomycin resistance could be 100% in ten years, but might also stay around 0%.</p>
|
||||
<p>You can define the model with the <code>model</code> parameter. The default model is a generalised linear regression model using a binomial distribution, assuming that a period of zero resistance was followed by a period of increasing resistance leading slowly to more and more resistance.</p>
|
||||
@ -346,25 +339,25 @@
|
||||
</tbody>
|
||||
</table>
|
||||
<p>For the vancomycin resistance in Gram positive bacteria, a linear model might be more appropriate since no (left half of a) binomial distribution is to be expected based on the observed years:</p>
|
||||
<div class="sourceCode" id="cb9"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb9-1" title="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb9-2" title="2"><span class="st"> </span><span class="kw"><a href="https://dplyr.tidyverse.org/reference/filter.html">filter</a></span>(<span class="kw"><a href="../reference/mo_property.html">mo_gramstain</a></span>(mo, <span class="dt">language =</span> <span class="ot">NULL</span>) <span class="op">==</span><span class="st"> "Gram-positive"</span>) <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb9-3" title="3"><span class="st"> </span><span class="kw"><a href="../reference/resistance_predict.html">resistance_predict</a></span>(<span class="dt">col_ab =</span> <span class="st">"VAN"</span>, <span class="dt">year_min =</span> <span class="dv">2010</span>, <span class="dt">info =</span> <span class="ot">FALSE</span>, <span class="dt">model =</span> <span class="st">"linear"</span>) <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb9-4" title="4"><span class="st"> </span><span class="kw"><a href="../reference/resistance_predict.html">ggplot_rsi_predict</a></span>()</a>
|
||||
<a class="sourceLine" id="cb9-5" title="5"><span class="co"># </span><span class="al">NOTE</span><span class="co">: Using column `date` as input for `col_date`.</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb9"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb9-1" data-line-number="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb9-2" data-line-number="2"><span class="st"> </span><span class="kw"><a href="https://dplyr.tidyverse.org/reference/filter.html">filter</a></span>(<span class="kw"><a href="../reference/mo_property.html">mo_gramstain</a></span>(mo, <span class="dt">language =</span> <span class="ot">NULL</span>) <span class="op">==</span><span class="st"> "Gram-positive"</span>) <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb9-3" data-line-number="3"><span class="st"> </span><span class="kw"><a href="../reference/resistance_predict.html">resistance_predict</a></span>(<span class="dt">col_ab =</span> <span class="st">"VAN"</span>, <span class="dt">year_min =</span> <span class="dv">2010</span>, <span class="dt">info =</span> <span class="ot">FALSE</span>, <span class="dt">model =</span> <span class="st">"linear"</span>) <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb9-4" data-line-number="4"><span class="st"> </span><span class="kw"><a href="../reference/resistance_predict.html">ggplot_rsi_predict</a></span>()</a>
|
||||
<a class="sourceLine" id="cb9-5" data-line-number="5"><span class="co"># </span><span class="al">NOTE</span><span class="co">: Using column `date` as input for `col_date`.</span></a></code></pre></div>
|
||||
<p><img src="resistance_predict_files/figure-html/unnamed-chunk-7-1.png" width="720"></p>
|
||||
<p>This seems more likely, doesn’t it?</p>
|
||||
<p>The model itself is also available from the object, as an <code>attribute</code>:</p>
|
||||
<div class="sourceCode" id="cb10"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb10-1" title="1">model <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/attributes">attributes</a></span>(predict_TZP)<span class="op">$</span>model</a>
|
||||
<a class="sourceLine" id="cb10-2" title="2"></a>
|
||||
<a class="sourceLine" id="cb10-3" title="3"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/summary">summary</a></span>(model)<span class="op">$</span>family</a>
|
||||
<a class="sourceLine" id="cb10-4" title="4"><span class="co"># </span></a>
|
||||
<a class="sourceLine" id="cb10-5" title="5"><span class="co"># Family: binomial </span></a>
|
||||
<a class="sourceLine" id="cb10-6" title="6"><span class="co"># Link function: logit</span></a>
|
||||
<a class="sourceLine" id="cb10-7" title="7"></a>
|
||||
<a class="sourceLine" id="cb10-8" title="8"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/summary">summary</a></span>(model)<span class="op">$</span>coefficients</a>
|
||||
<a class="sourceLine" id="cb10-9" title="9"><span class="co"># Estimate Std. Error z value Pr(>|z|)</span></a>
|
||||
<a class="sourceLine" id="cb10-10" title="10"><span class="co"># (Intercept) -224.3987194 48.0335384 -4.671709 2.987038e-06</span></a>
|
||||
<a class="sourceLine" id="cb10-11" title="11"><span class="co"># year 0.1106102 0.0238753 4.632831 3.606990e-06</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb10"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb10-1" data-line-number="1">model <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/attributes">attributes</a></span>(predict_TZP)<span class="op">$</span>model</a>
|
||||
<a class="sourceLine" id="cb10-2" data-line-number="2"></a>
|
||||
<a class="sourceLine" id="cb10-3" data-line-number="3"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/summary">summary</a></span>(model)<span class="op">$</span>family</a>
|
||||
<a class="sourceLine" id="cb10-4" data-line-number="4"><span class="co"># </span></a>
|
||||
<a class="sourceLine" id="cb10-5" data-line-number="5"><span class="co"># Family: binomial </span></a>
|
||||
<a class="sourceLine" id="cb10-6" data-line-number="6"><span class="co"># Link function: logit</span></a>
|
||||
<a class="sourceLine" id="cb10-7" data-line-number="7"></a>
|
||||
<a class="sourceLine" id="cb10-8" data-line-number="8"><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/summary">summary</a></span>(model)<span class="op">$</span>coefficients</a>
|
||||
<a class="sourceLine" id="cb10-9" data-line-number="9"><span class="co"># Estimate Std. Error z value Pr(>|z|)</span></a>
|
||||
<a class="sourceLine" id="cb10-10" data-line-number="10"><span class="co"># (Intercept) -224.3987194 48.0335384 -4.671709 2.987038e-06</span></a>
|
||||
<a class="sourceLine" id="cb10-11" data-line-number="11"><span class="co"># year 0.1106102 0.0238753 4.632831 3.606990e-06</span></a></code></pre></div>
|
||||
</div>
|
||||
</div>
|
||||
</div>
|
||||
|
||||
|
Before 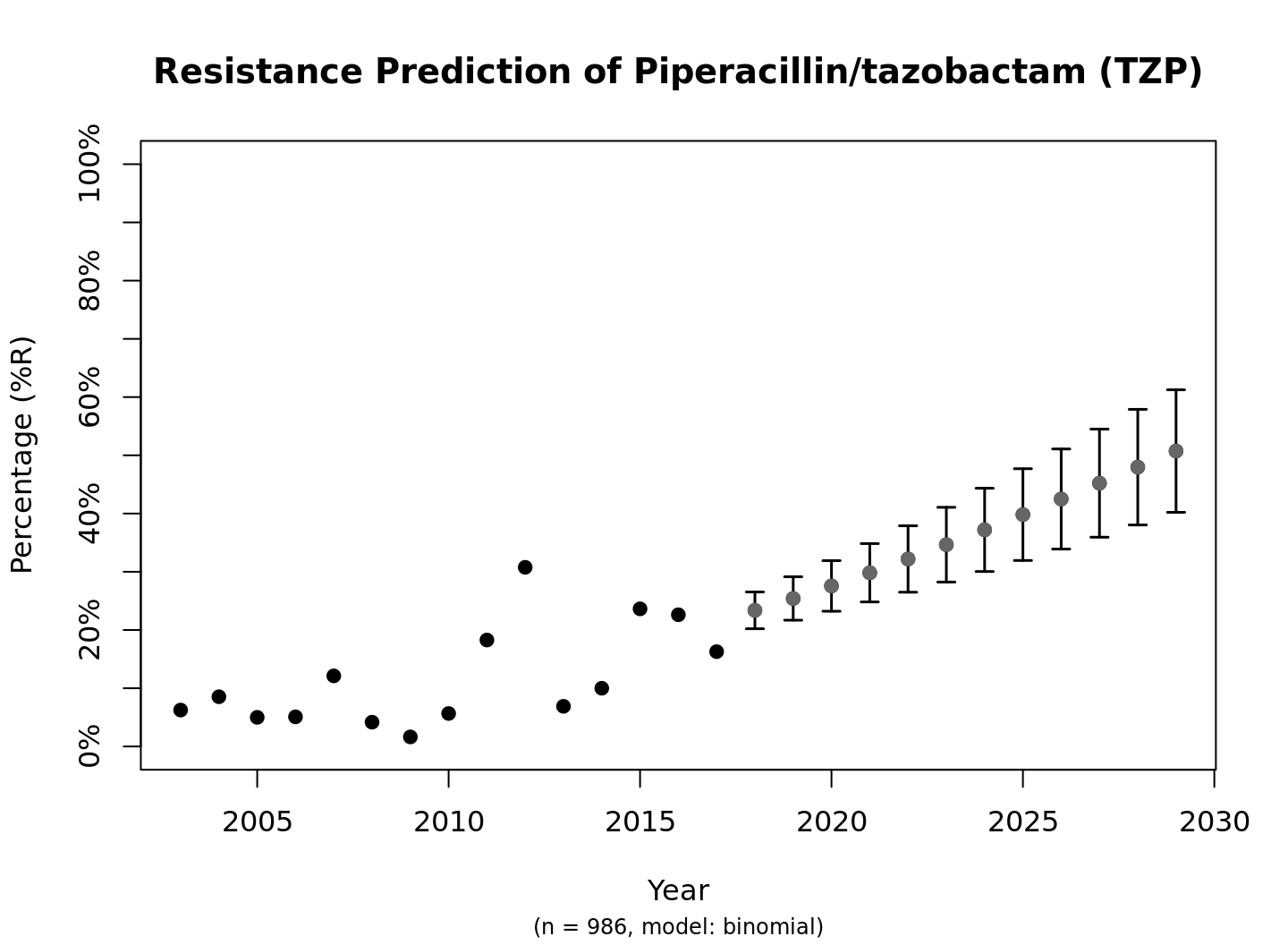
(image error) Size: 30 KiB After 
(image error) Size: 95 KiB 

|
|
Before 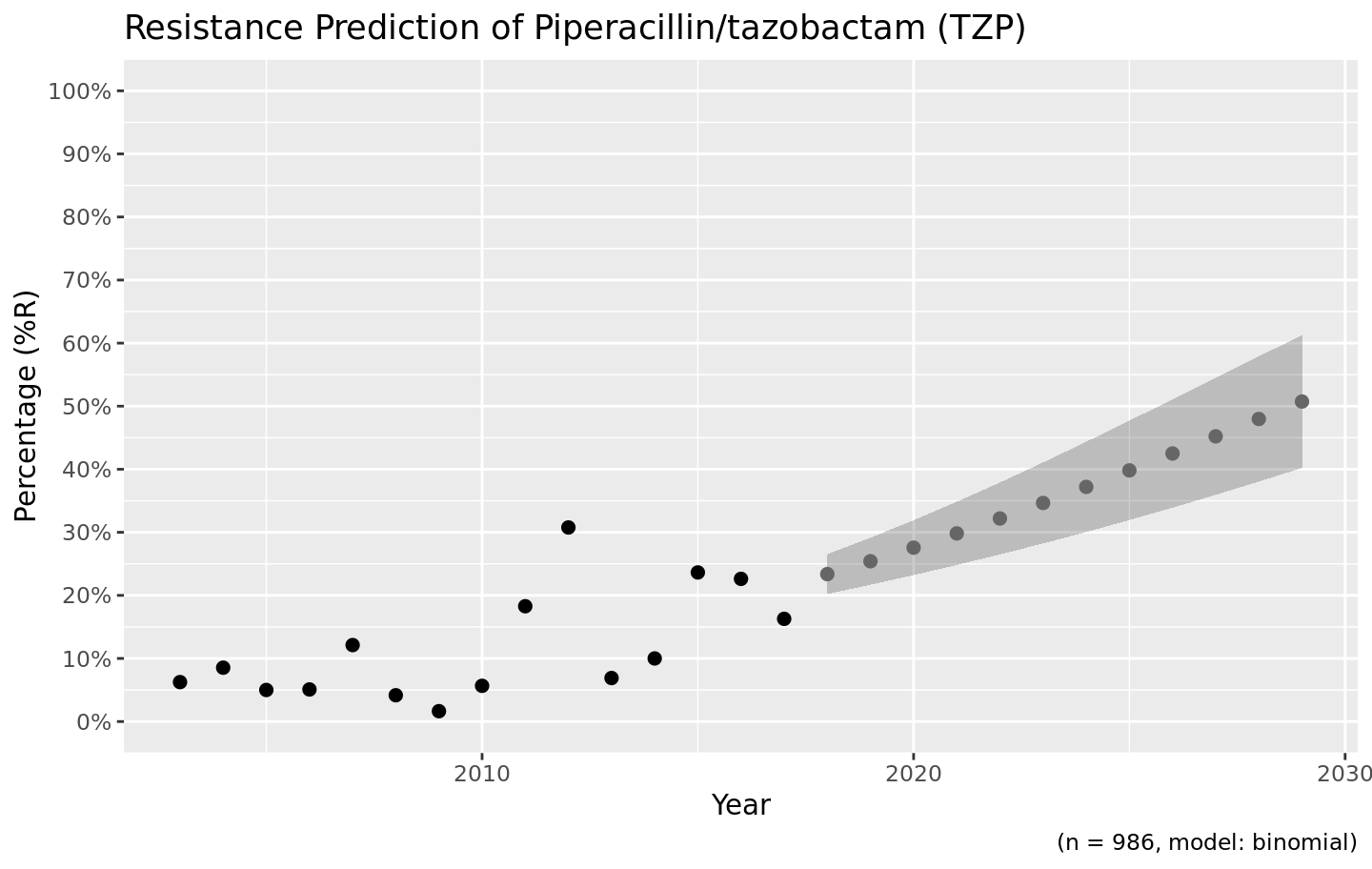
(image error) Size: 32 KiB After 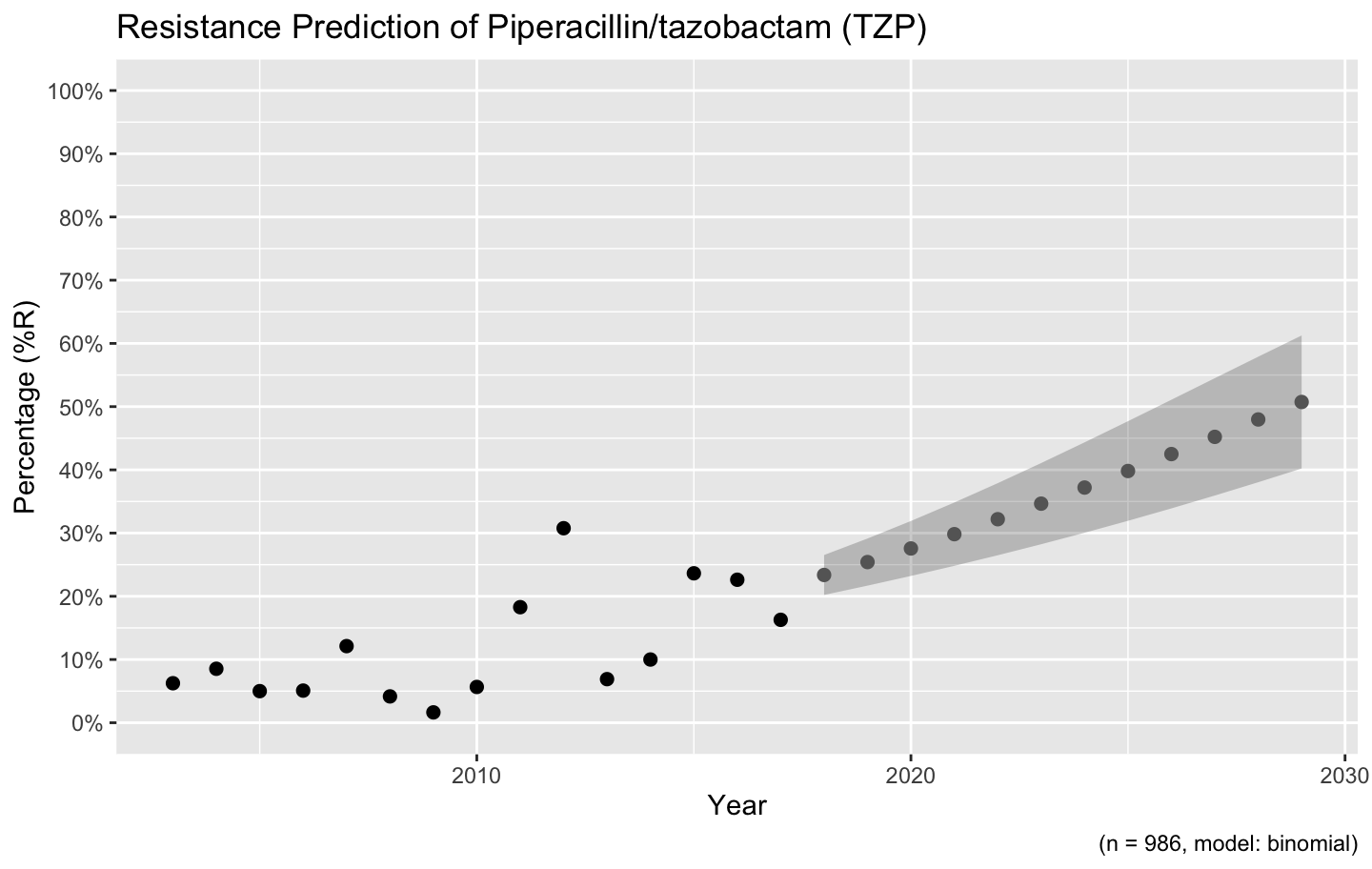
(image error) Size: 94 KiB 

|
|
Before 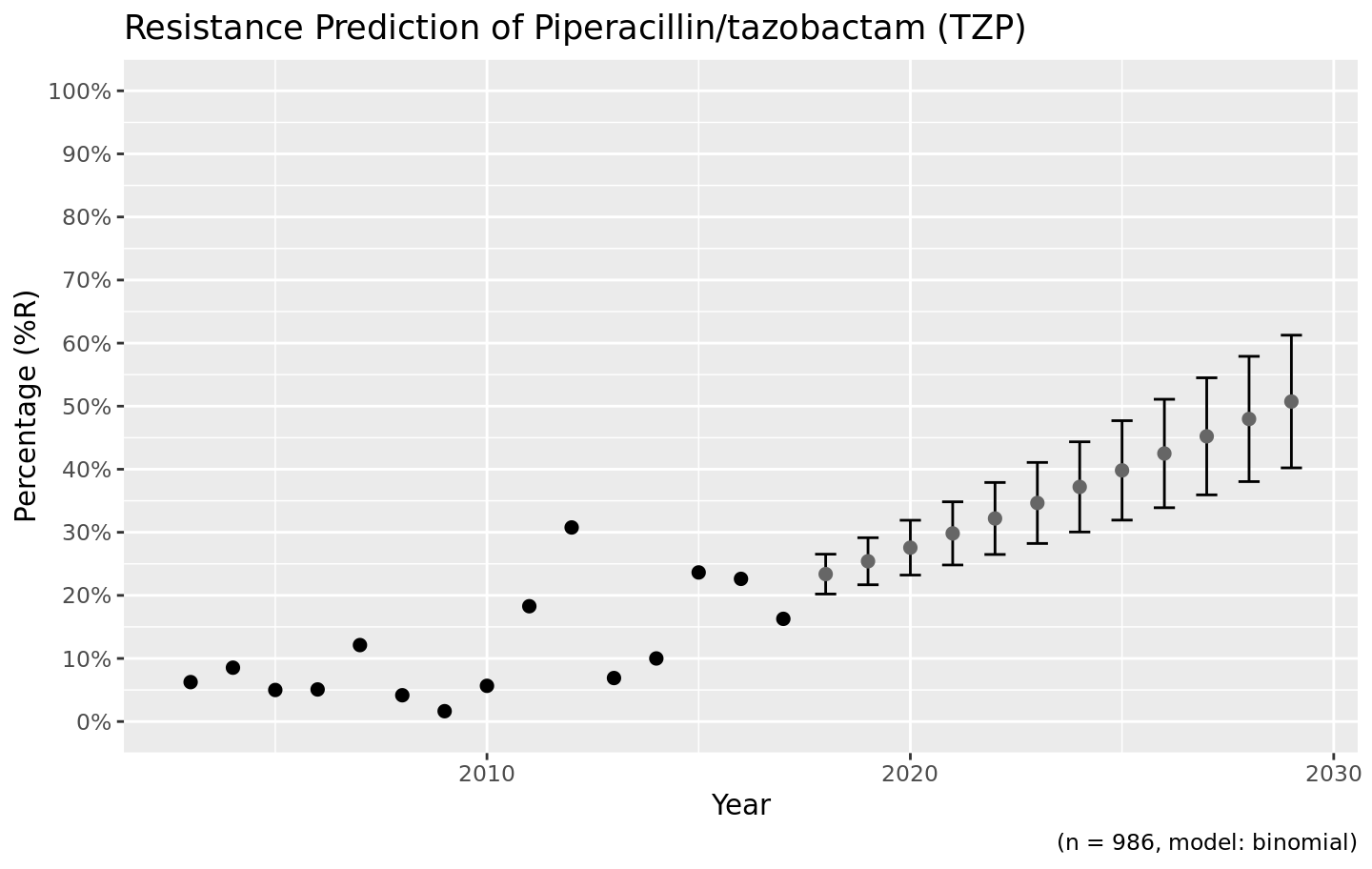
(image error) Size: 32 KiB After 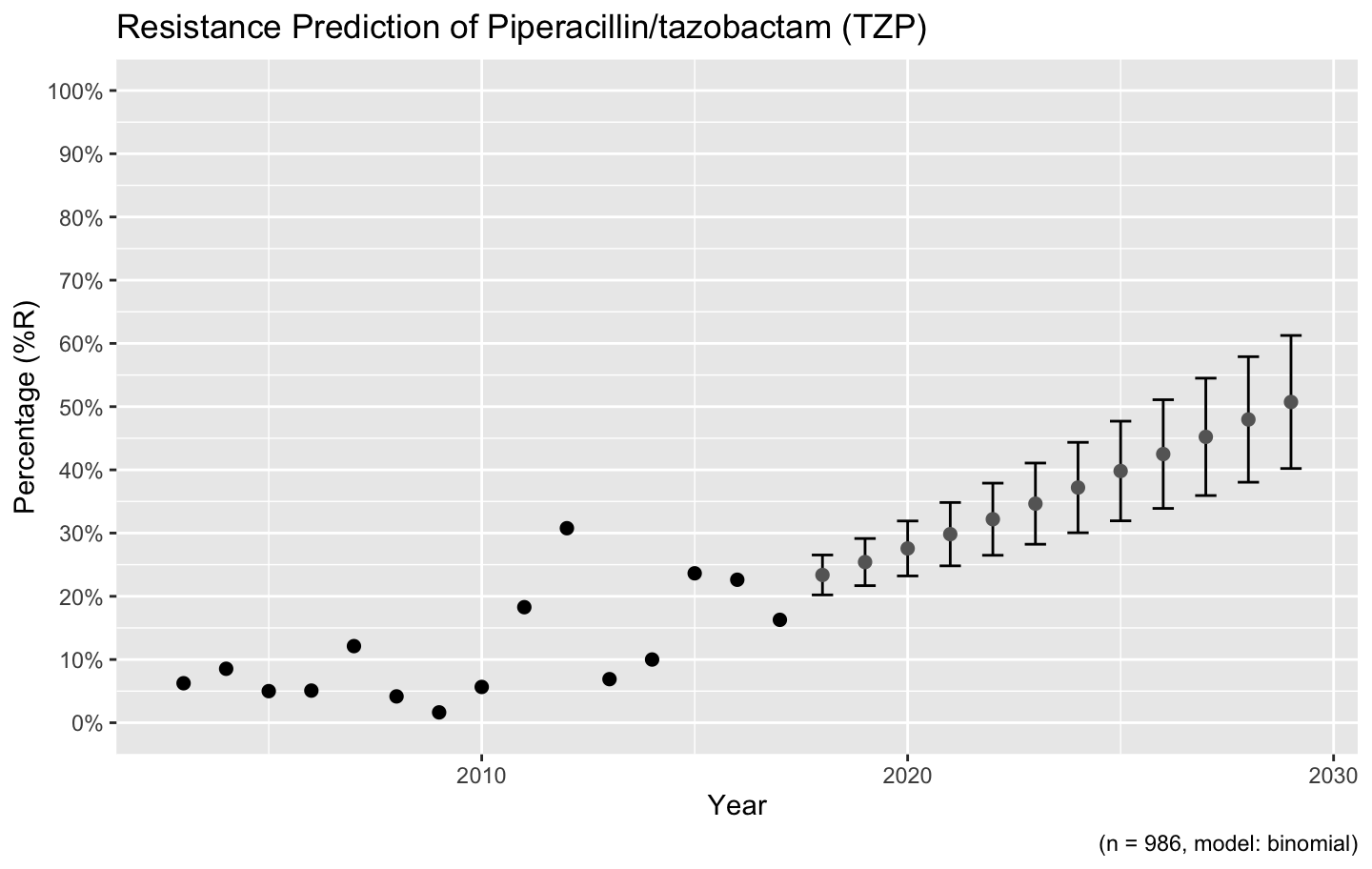
(image error) Size: 91 KiB 

|
|
Before 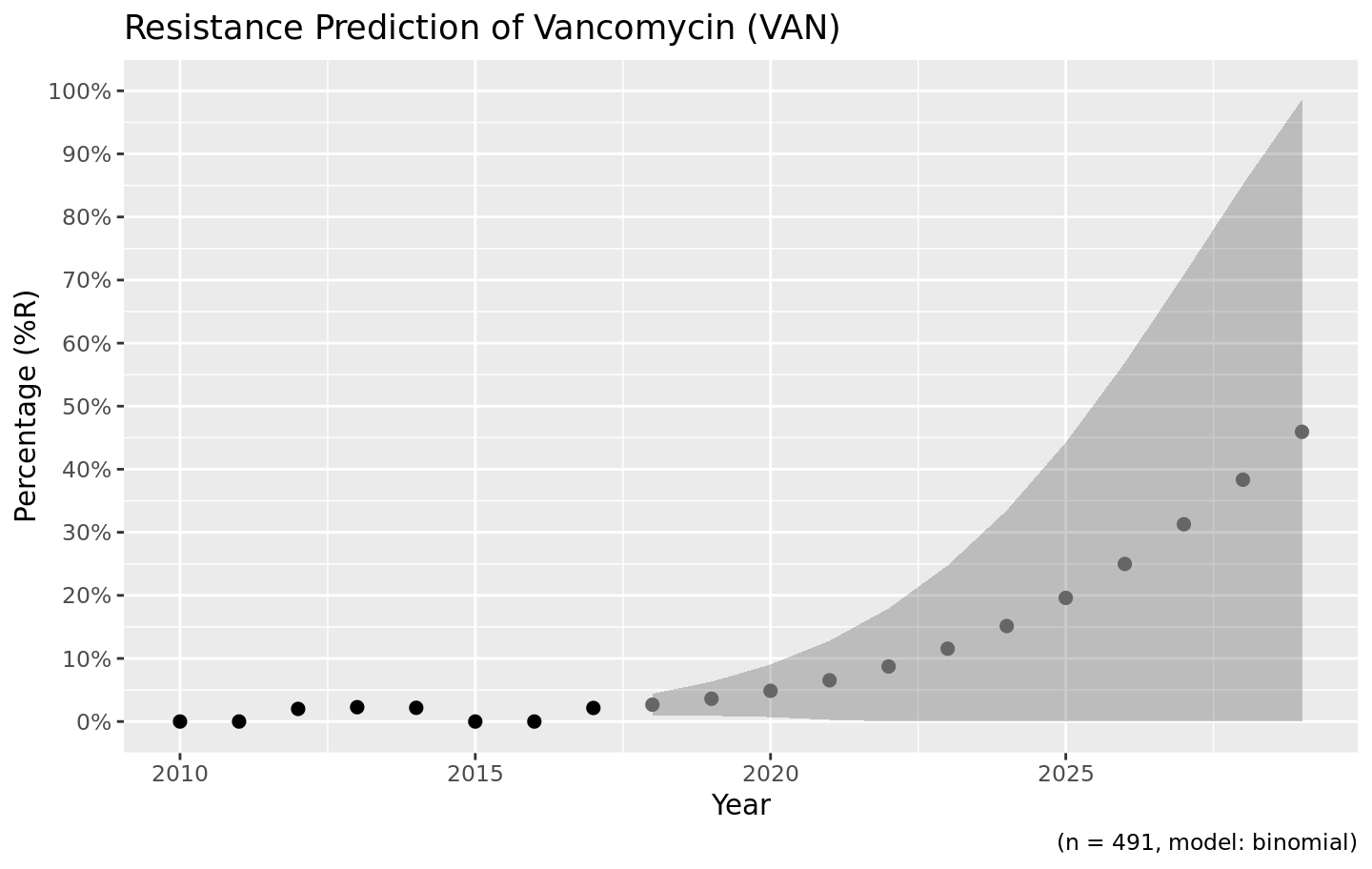
(image error) Size: 34 KiB After 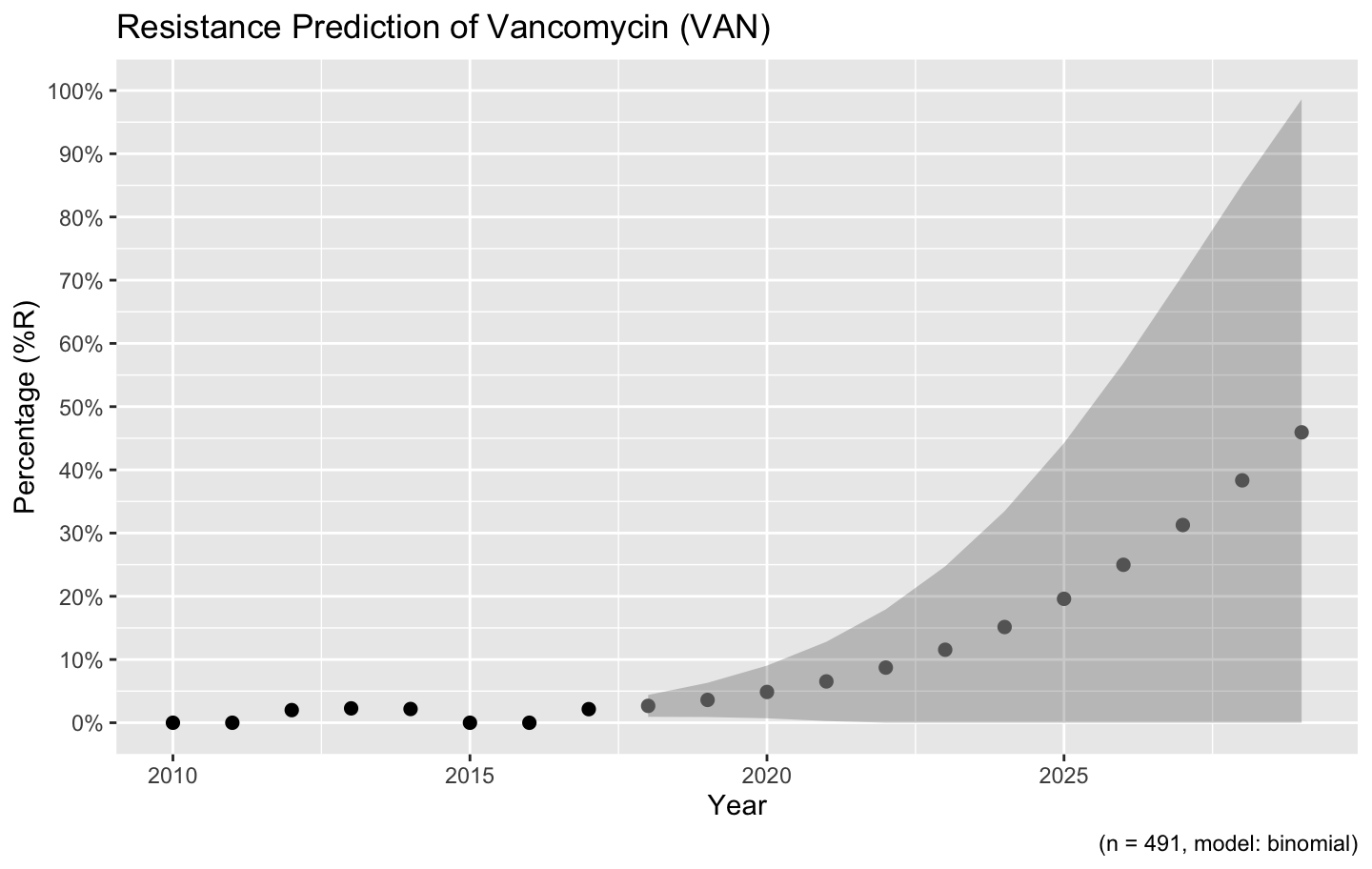
(image error) Size: 94 KiB 

|
|
Before 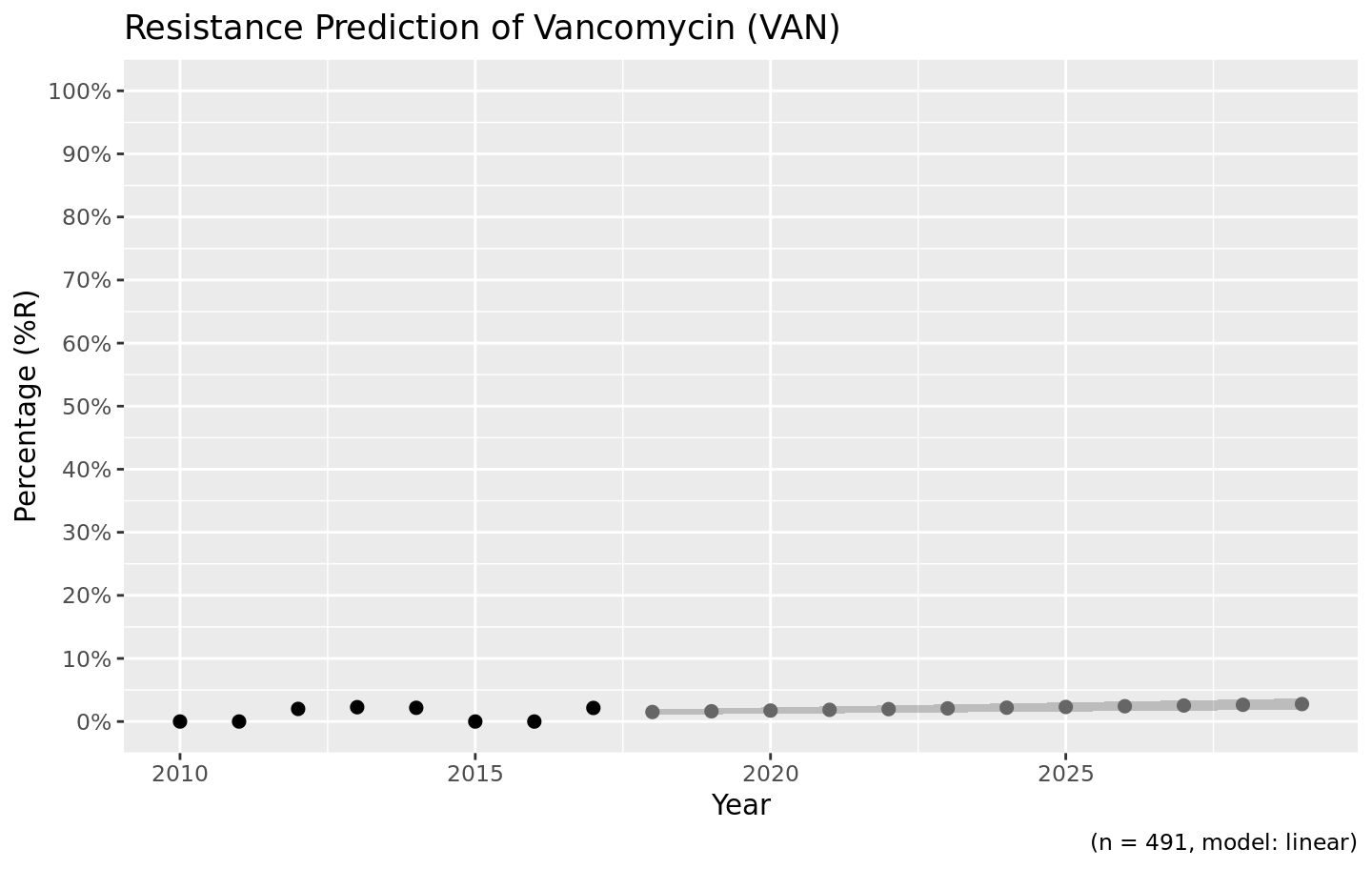
(image error) Size: 31 KiB After 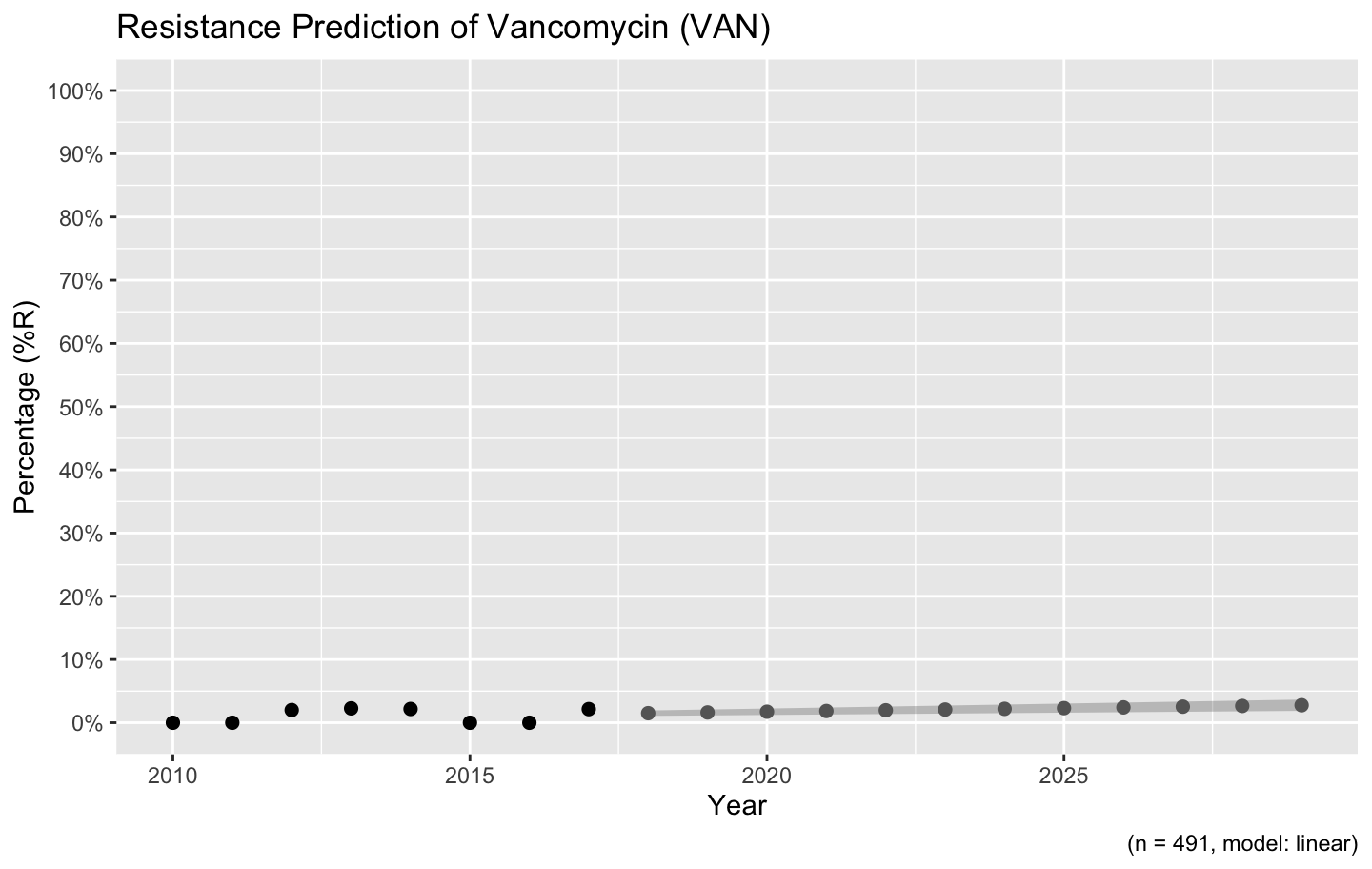
(image error) Size: 87 KiB 

|
@ -78,7 +78,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9008</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9026</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -156,13 +156,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -187,8 +187,10 @@ table a:not(.btn):hover, .table a:not(.btn):hover {
|
||||
|
||||
/* text below header in manual overview */
|
||||
.template-reference-index h2 ~ p {
|
||||
font-size: 110%;
|
||||
/* font-weight: bold; */
|
||||
font-size: 16px;
|
||||
}
|
||||
.template-reference-topic h2 {
|
||||
font-size: 24px;
|
||||
}
|
||||
|
||||
/* logos on index page */
|
||||
|
||||
@ -46,6 +46,9 @@ $( document ).ready(function() {
|
||||
window.location.replace(url_new);
|
||||
}
|
||||
|
||||
// Replace 'Value' in manual to 'Returned value'
|
||||
$(".template-reference-topic h2#value").text("Returned value");
|
||||
|
||||
// PR for 'R for Data Science' on How To pages
|
||||
if ($(".template-article").length > 0) {
|
||||
$('#sidebar').prepend(
|
||||
|
||||
@ -42,7 +42,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9008</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9026</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -120,13 +120,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -190,9 +183,9 @@
|
||||
|
||||
</header><div class="row">
|
||||
<div class="contents col-md-9">
|
||||
<div id="amr-for-r-" class="section level1">
|
||||
<div id="amr-for-r" class="section level1">
|
||||
<div class="page-header"><h1 class="hasAnchor">
|
||||
<a href="#amr-for-r-" class="anchor"></a><code>AMR</code> (for R) <img src="./logo.png" align="right" height="120px">
|
||||
<a href="#amr-for-r" class="anchor"></a><code>AMR</code> (for R) <img src="./logo.png" align="right" height="120px">
|
||||
</h1></div>
|
||||
<p><em>(<help title="Too Long, Didn't Read">TLDR</help> - to find out how to conduct AMR analysis, please <a href="./articles/AMR.html">continue reading here to get started</a>.</em></p>
|
||||
<hr>
|
||||
@ -204,16 +197,15 @@
|
||||
<ul>
|
||||
<li>Reference for the taxonomy of microorganisms, since the package contains all microbial (sub)species from the <a href="http://www.catalogueoflife.org">Catalogue of Life</a> (<a href="./reference/mo_property.html">manual</a>)</li>
|
||||
<li>Interpreting raw MIC and disk diffusion values, based on the latest CLSI or EUCAST guidelines (<a href="./reference/as.rsi.html">manual</a>)</li>
|
||||
<li>Determining first isolates to be used for AMR analysis (<a href="./reference/first_isolate.html">manual</a>)</li>
|
||||
<li>Calculating antimicrobial resistance (<a href="./articles/AMR.html">tutorial</a>)</li>
|
||||
<li>Determining multi-drug resistance (MDR) / multi-drug resistant organisms (MDRO) (<a href="./articles/MDR.html">tutorial</a>)</li>
|
||||
<li>Calculating empirical susceptibility of both mono therapy and combination therapy (<a href="./articles/AMR.html">tutorial</a>)</li>
|
||||
<li>Calculating (empirical) susceptibility of both mono therapy and combination therapies (<a href="./articles/AMR.html">tutorial</a>)</li>
|
||||
<li>Predicting future antimicrobial resistance using regression models (<a href="./articles/resistance_predict.html">tutorial</a>)</li>
|
||||
<li>Getting properties for any microorganism (like Gram stain, species, genus or family) (<a href="./reference/mo_property.html">manual</a>)</li>
|
||||
<li>Getting properties for any antibiotic (like name, ATC code, defined daily dose or trade name) (<a href="./reference/ab_property.html">manual</a>)</li>
|
||||
<li>Plotting antimicrobial resistance (<a href="./articles/AMR.html">tutorial</a>)</li>
|
||||
<li>Determining first isolates to be used for AMR analysis (<a href="./reference/first_isolate.html">manual</a>)</li>
|
||||
<li>Applying EUCAST expert rules (<a href="./reference/eucast_rules.html">manual</a>)</li>
|
||||
<li>Descriptive statistics: frequency tables, kurtosis and skewness (<a href="./articles/freq.html">tutorial</a>)</li>
|
||||
</ul>
|
||||
<p>This package is ready-to-use for a professional environment by specialists in the following fields:</p>
|
||||
<p>Medical Microbiology</p>
|
||||
@ -248,7 +240,7 @@
|
||||
<h4 class="hasAnchor">
|
||||
<a href="#latest-released-version" class="anchor"></a>Latest released version</h4>
|
||||
<p>This package is available <a href="https://cran.r-project.org/package=AMR">on the official R network (CRAN)</a>, which has a peer-reviewed submission process. Install this package in R with:</p>
|
||||
<div class="sourceCode" id="cb1"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb1-1" title="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/utils/topics/install.packages">install.packages</a></span>(<span class="st">"AMR"</span>)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb1"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb1-1" data-line-number="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/utils/topics/install.packages">install.packages</a></span>(<span class="st">"AMR"</span>)</a></code></pre></div>
|
||||
<p>It will be downloaded and installed automatically. For RStudio, click on the menu <em>Tools</em> > <em>Install Packages…</em> and then type in “AMR” and press <kbd>Install</kbd>.</p>
|
||||
<p><strong>Note:</strong> Not all functions on this website may be available in this latest release. To use all functions and data sets mentioned on this website, install the latest development version.</p>
|
||||
</div>
|
||||
@ -256,8 +248,8 @@
|
||||
<h4 class="hasAnchor">
|
||||
<a href="#latest-development-version" class="anchor"></a>Latest development version</h4>
|
||||
<p>The latest and unpublished development version can be installed with (<strong>precaution: may be unstable</strong>):</p>
|
||||
<div class="sourceCode" id="cb2"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb2-1" title="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/utils/topics/install.packages">install.packages</a></span>(<span class="st">"devtools"</span>)</a>
|
||||
<a class="sourceLine" id="cb2-2" title="2">devtools<span class="op">::</span><span class="kw"><a href="https://www.rdocumentation.org/packages/devtools/topics/reexports">install_gitlab</a></span>(<span class="st">"msberends/AMR"</span>)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb2"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb2-1" data-line-number="1"><span class="kw"><a href="https://www.rdocumentation.org/packages/utils/topics/install.packages">install.packages</a></span>(<span class="st">"devtools"</span>)</a>
|
||||
<a class="sourceLine" id="cb2-2" data-line-number="2">devtools<span class="op">::</span><span class="kw"><a href="https://www.rdocumentation.org/packages/devtools/topics/reexports">install_gitlab</a></span>(<span class="st">"msberends/AMR"</span>)</a></code></pre></div>
|
||||
</div>
|
||||
</div>
|
||||
<div id="get-started" class="section level2">
|
||||
@ -296,11 +288,12 @@
|
||||
<img src="reference/figures/logo_who.png" height="75px" class="logo_img"><p class="logo_txt">WHO Collaborating Centre for Drug Statistics Methodology</p>
|
||||
</div>
|
||||
<p>This package contains <strong>all ~450 antimicrobial drugs</strong> and their Anatomical Therapeutic Chemical (ATC) codes, ATC groups and Defined Daily Dose (DDD, oral and IV) from the World Health Organization Collaborating Centre for Drug Statistics Methodology (WHOCC, <a href="https://www.whocc.no" class="uri">https://www.whocc.no</a>) and the <a href="http://ec.europa.eu/health/documents/community-register/html/atc.htm">Pharmaceuticals Community Register of the European Commission</a>.</p>
|
||||
<p><strong>NOTE: The WHOCC copyright does not allow use for commercial purposes, unlike any other info from this package. See \url{<a href="https://www.whocc.no/copyright_disclaimer/" class="uri">https://www.whocc.no/copyright_disclaimer/</a>}.</strong></p>
|
||||
<p>Read more about the data from WHOCC <a href="./reference/WHOCC.html">in our manual</a>.</p>
|
||||
</div>
|
||||
<div id="whonet--ears-net" class="section level4">
|
||||
<div id="whonet-ears-net" class="section level4">
|
||||
<h4 class="hasAnchor">
|
||||
<a href="#whonet--ears-net" class="anchor"></a>WHONET / EARS-Net</h4>
|
||||
<a href="#whonet-ears-net" class="anchor"></a>WHONET / EARS-Net</h4>
|
||||
<p><img src="./whonet.png"></p>
|
||||
<p>We support WHONET and EARS-Net data. Exported files from WHONET can be imported into R and can be analysed easily using this package. For education purposes, we created an <a href="./reference/WHONET.html">example data set <code>WHONET</code></a> with the exact same structure as a WHONET export file. Furthermore, this package also contains a <a href="./reference/antibiotics.html">data set <code>antibiotics</code></a> with all EARS-Net antibiotic abbreviations, and knows almost all WHONET abbreviations for microorganisms. When using WHONET data as input for analysis, all input parameters will be set automatically.</p>
|
||||
<p>Read our tutorial about <a href="./articles/WHONET.html">how to work with WHONET data here</a>.</p>
|
||||
@ -314,7 +307,7 @@
|
||||
<p>It <strong>cleanses existing data</strong> by providing new <em>classes</em> for microoganisms, antibiotics and antimicrobial results (both S/I/R and MIC). By installing this package, you teach R everything about microbiology that is needed for analysis. These functions all use intelligent rules to guess results that you would expect:</p>
|
||||
<ul>
|
||||
<li>Use <code><a href="reference/as.mo.html">as.mo()</a></code> to get a microbial ID. The IDs are human readable for the trained eye - the ID of <em>Klebsiella pneumoniae</em> is “B_KLBSL_PNE” (B stands for Bacteria) and the ID of <em>S. aureus</em> is “B_STPHY_AUR”. The function takes almost any text as input that looks like the name or code of a microorganism like “E. coli”, “esco” or “esccol” and tries to find expected results using intelligent rules combined with the included Catalogue of Life data set. It only takes milliseconds to find results, please see our <a href="./articles/benchmarks.html">benchmarks</a>. Moreover, it can group <em>Staphylococci</em> into coagulase negative and positive (CoNS and CoPS, see <a href="./reference/as.mo.html#source">source</a>) and can categorise <em>Streptococci</em> into Lancefield groups (like beta-haemolytic <em>Streptococcus</em> Group B, <a href="./reference/as.mo.html#source">source</a>).</li>
|
||||
<li>Use <code><a href="reference/as.ab.html">as.ab()</a></code> to get an antibiotic ID. Like microbial IDs, these IDs are also human readable based on those used by EARS-Net. For example, the ID of amoxicillin is <code>AMX</code> and the ID of gentamicin is <code>GEN</code>. The <code><a href="reference/as.ab.html">as.ab()</a></code> function also uses intelligent rules to find results like accepting misspelling, trade names and abbrevations used in many laboratory systems. For instance, the values “Furabid”, “Furadantin”, “nitro” all return the ID of Nitrofurantoine. To accomplish this, the package contains a database with most LIS codes, official names, trade names, DDDs and categories of antibiotics. The function <code><a href="reference/AMR-deprecated.html">as.atc()</a></code> will return the ATC code of an antibiotic as defined by the WHO.</li>
|
||||
<li>Use <code><a href="reference/as.ab.html">as.ab()</a></code> to get an antibiotic ID. Like microbial IDs, these IDs are also human readable based on those used by EARS-Net. For example, the ID of amoxicillin is <code>AMX</code> and the ID of gentamicin is <code>GEN</code>. The <code><a href="reference/as.ab.html">as.ab()</a></code> function also uses intelligent rules to find results like accepting misspelling, trade names and abbrevations used in many laboratory systems. For instance, the values “Furabid”, “Furadantin”, “nitro” all return the ID of Nitrofurantoine. To accomplish this, the package contains a database with most LIS codes, official names, trade names, ATC codes, defined daily doses (DDD) and drug categories of antibiotics.</li>
|
||||
<li>Use <code><a href="reference/as.rsi.html">as.rsi()</a></code> to get antibiotic interpretations based on raw MIC values (in mg/L) or disk diffusion values (in mm), or transform existing values to valid antimicrobial results. It produces just S, I or R based on your input and warns about invalid values. Even values like “<=0.002; S” (combined MIC/RSI) will result in “S”.</li>
|
||||
<li>Use <code><a href="reference/as.mic.html">as.mic()</a></code> to cleanse your MIC values. It produces a so-called factor (called <em>ordinal</em> in SPSS) with valid MIC values as levels. A value like “<=0.002; S” (combined MIC/RSI) will result in “<=0.002”.</li>
|
||||
</ul>
|
||||
@ -330,7 +323,7 @@
|
||||
</li>
|
||||
<li>Use <code><a href="reference/mdro.html">mdro()</a></code> (abbreviation of Multi Drug Resistant Organisms) to check your isolates for exceptional resistance with country-specific guidelines or EUCAST rules. Currently, national guidelines for Germany and the Netherlands are supported.</li>
|
||||
<li>The <a href="./reference/microorganisms.html">data set <code>microorganisms</code></a> contains the complete taxonomic tree of ~65,000 microorganisms. Furthermore, some colloquial names and all Gram stains are available, which enables resistance analysis of e.g. different antibiotics per Gram stain. The package also contains functions to look up values in this data set like <code><a href="reference/mo_property.html">mo_genus()</a></code>, <code><a href="reference/mo_property.html">mo_family()</a></code>, <code><a href="reference/mo_property.html">mo_gramstain()</a></code> or even <code><a href="reference/mo_property.html">mo_phylum()</a></code>. As they use <code><a href="reference/as.mo.html">as.mo()</a></code> internally, they also use the same intelligent rules for determination. For example, <code><a href="reference/mo_property.html">mo_genus("MRSA")</a></code> and <code><a href="reference/mo_property.html">mo_genus("S. aureus")</a></code> will both return <code>"Staphylococcus"</code>. They also come with support for German, Dutch, Spanish, Italian, French and Portuguese. These functions can be used to add new variables to your data.</li>
|
||||
<li>The <a href="./reference/antibiotics.html">data set <code>antibiotics</code></a> contains ~450 antimicrobial drugs with their EARS-Net code, ATC code, PubChem compound ID, official name, common LIS codes and DDDs of both oral and parenteral administration. It also contains all (thousands of) trade names found in PubChem. Use functions like <code><a href="reference/ab_property.html">ab_name()</a></code>, <code><a href="reference/ab_property.html">ab_group()</a></code> and <code><a href="reference/ab_property.html">ab_tradenames()</a></code> to look up values. The <code>ab_*</code> functions use <code><a href="reference/as.ab.html">as.ab()</a></code> internally so they support the same intelligent rules to guess the most probable result. For example, <code><a href="reference/ab_property.html">ab_name("Fluclox")</a></code>, <code><a href="reference/ab_property.html">ab_name("Floxapen")</a></code> and <code><a href="reference/ab_property.html">ab_name("J01CF05")</a></code> will all return <code>"Flucloxacillin"</code>. These functions can again be used to add new variables to your data.</li>
|
||||
<li>The <a href="./reference/antibiotics.html">data set <code>antibiotics</code></a> contains ~450 antimicrobial drugs with their EARS-Net code, ATC code, PubChem compound ID, official name, common LIS codes and DDDs of both oral and parenteral administration. It also contains all (thousands of) trade names found in PubChem. The function <code><a href="reference/ab_property.html">ab_atc()</a></code> will return the ATC code of an antibiotic as defined by the WHO. Use functions like <code><a href="reference/ab_property.html">ab_name()</a></code>, <code><a href="reference/ab_property.html">ab_group()</a></code> and <code><a href="reference/ab_property.html">ab_tradenames()</a></code> to look up values. The <code>ab_*</code> functions use <code><a href="reference/as.ab.html">as.ab()</a></code> internally so they support the same intelligent rules to guess the most probable result. For example, <code><a href="reference/ab_property.html">ab_name("Fluclox")</a></code>, <code><a href="reference/ab_property.html">ab_name("Floxapen")</a></code> and <code><a href="reference/ab_property.html">ab_name("J01CF05")</a></code> will all return <code>"Flucloxacillin"</code>. These functions can again be used to add new variables to your data.</li>
|
||||
</ul>
|
||||
</li>
|
||||
<li>
|
||||
@ -339,8 +332,6 @@
|
||||
<li>Calculate the resistance (and even co-resistance) of microbial isolates with the <code><a href="reference/portion.html">portion_R()</a></code>, <code><a href="reference/portion.html">portion_IR()</a></code>, <code><a href="reference/portion.html">portion_I()</a></code>, <code><a href="reference/portion.html">portion_SI()</a></code> and <code><a href="reference/portion.html">portion_S()</a></code> functions. Similarly, the <em>number</em> of isolates can be determined with the <code><a href="reference/count.html">count_R()</a></code>, <code><a href="reference/count.html">count_IR()</a></code>, <code><a href="reference/count.html">count_I()</a></code>, <code><a href="reference/count.html">count_SI()</a></code> and <code><a href="reference/count.html">count_S()</a></code> functions. All these functions can be used with the <code>dplyr</code> package (e.g. in conjunction with <code>summarise()</code>)</li>
|
||||
<li>Plot AMR results with <code><a href="reference/ggplot_rsi.html">geom_rsi()</a></code>, a function made for the <code>ggplot2</code> package</li>
|
||||
<li>Predict antimicrobial resistance for the nextcoming years using logistic regression models with the <code><a href="reference/resistance_predict.html">resistance_predict()</a></code> function</li>
|
||||
<li>Conduct descriptive statistics to enhance base R: calculate <code><a href="reference/kurtosis.html">kurtosis()</a></code>, <code><a href="reference/skewness.html">skewness()</a></code> and create frequency tables with <code><a href="reference/freq.html">freq()</a></code>
|
||||
</li>
|
||||
</ul>
|
||||
</li>
|
||||
<li>
|
||||
@ -386,9 +377,7 @@
|
||||
<div class="license">
|
||||
<h2>License</h2>
|
||||
<ul class="list-unstyled">
|
||||
<li>
|
||||
<a href="https://www.r-project.org/Licenses/GPL-2">GPL-2</a> | file <a href="LICENSE-text.html">LICENSE</a>
|
||||
</li>
|
||||
<li><a href="https://www.r-project.org/Licenses/GPL-2">GPL-2</a></li>
|
||||
</ul>
|
||||
</div>
|
||||
<div class="developers">
|
||||
|
||||
@ -78,7 +78,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9008</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9026</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -156,13 +156,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -232,39 +225,46 @@
|
||||
|
||||
</div>
|
||||
|
||||
<div id="amr-0719008" class="section level1">
|
||||
<div id="amr-0-7-1-9026" class="section level1">
|
||||
<h1 class="page-header">
|
||||
<a href="#amr-0719008" class="anchor"></a>AMR 0.7.1.9008<small> Unreleased </small>
|
||||
<a href="#amr-0-7-1-9026" class="anchor"></a>AMR 0.7.1.9026<small> Unreleased </small>
|
||||
</h1>
|
||||
<div id="breaking" class="section level3">
|
||||
<h3 class="hasAnchor">
|
||||
<a href="#breaking" class="anchor"></a>Breaking</h3>
|
||||
<ul>
|
||||
<li>Function <code>freq()</code> has moved to a new package, <a href="https://github.com/msberends/clean"><code>clean</code></a> (<a href="https://cran.r-project.org/package=clean">CRAN link</a>). Creating frequency tables is actually not the scope of this package (never was) and this function has matured a lot over the last two years. Therefore, a new package was created for data cleaning and checking and it perfectly fits the <code>freq()</code> function. The <a href="https://github.com/msberends/clean"><code>clean</code></a> package is available on CRAN and will be installed automatically when updating the <code>AMR</code> package, that now imports it. In a later stage, the <code><a href="../reference/skewness.html">skewness()</a></code> and <code><a href="../reference/kurtosis.html">kurtosis()</a></code> functions will be moved to the <code>clean</code> package too.</li>
|
||||
</ul>
|
||||
</div>
|
||||
<div id="new" class="section level3">
|
||||
<h3 class="hasAnchor">
|
||||
<a href="#new" class="anchor"></a>New</h3>
|
||||
<ul>
|
||||
<li>
|
||||
<p>Additional way to calculate co-resistance, i.e. when using multiple antibiotics as input for <code>portion_*</code> functions or <code>count_*</code> functions. This can be used to determine the empiric susceptibily of a combination therapy. A new parameter <code>only_all_tested</code> (<strong>which defaults to <code>FALSE</code></strong>) replaces the old <code>also_single_tested</code> and can be used to select one of the two methods to count isolates and calculate portions. The difference can be seen in this example table (which is also on the <code>portion</code> and <code>count</code> help pages), where the %SI is being determined:</p>
|
||||
<div class="sourceCode" id="cb1"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb1-1" title="1"><span class="co"># -------------------------------------------------------------------------</span></a>
|
||||
<a class="sourceLine" id="cb1-2" title="2"><span class="co"># only_all_tested = FALSE only_all_tested = TRUE</span></a>
|
||||
<a class="sourceLine" id="cb1-3" title="3"><span class="co"># Antibiotic Antibiotic ----------------------- -----------------------</span></a>
|
||||
<a class="sourceLine" id="cb1-4" title="4"><span class="co"># A B include as include as include as include as</span></a>
|
||||
<a class="sourceLine" id="cb1-5" title="5"><span class="co"># numerator denominator numerator denominator</span></a>
|
||||
<a class="sourceLine" id="cb1-6" title="6"><span class="co"># ---------- ---------- ---------- ----------- ---------- -----------</span></a>
|
||||
<a class="sourceLine" id="cb1-7" title="7"><span class="co"># S S X X X X</span></a>
|
||||
<a class="sourceLine" id="cb1-8" title="8"><span class="co"># I S X X X X</span></a>
|
||||
<a class="sourceLine" id="cb1-9" title="9"><span class="co"># R S X X X X</span></a>
|
||||
<a class="sourceLine" id="cb1-10" title="10"><span class="co"># not tested S X X - -</span></a>
|
||||
<a class="sourceLine" id="cb1-11" title="11"><span class="co"># S I X X X X</span></a>
|
||||
<a class="sourceLine" id="cb1-12" title="12"><span class="co"># I I X X X X</span></a>
|
||||
<a class="sourceLine" id="cb1-13" title="13"><span class="co"># R I X X X X</span></a>
|
||||
<a class="sourceLine" id="cb1-14" title="14"><span class="co"># not tested I X X - -</span></a>
|
||||
<a class="sourceLine" id="cb1-15" title="15"><span class="co"># S R X X X X</span></a>
|
||||
<a class="sourceLine" id="cb1-16" title="16"><span class="co"># I R X X X X</span></a>
|
||||
<a class="sourceLine" id="cb1-17" title="17"><span class="co"># R R - X - X</span></a>
|
||||
<a class="sourceLine" id="cb1-18" title="18"><span class="co"># not tested R - - - -</span></a>
|
||||
<a class="sourceLine" id="cb1-19" title="19"><span class="co"># S not tested X X - -</span></a>
|
||||
<a class="sourceLine" id="cb1-20" title="20"><span class="co"># I not tested X X - -</span></a>
|
||||
<a class="sourceLine" id="cb1-21" title="21"><span class="co"># R not tested - - - -</span></a>
|
||||
<a class="sourceLine" id="cb1-22" title="22"><span class="co"># not tested not tested - - - -</span></a>
|
||||
<a class="sourceLine" id="cb1-23" title="23"><span class="co"># -------------------------------------------------------------------------</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb1"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb1-1" data-line-number="1"><span class="co"># -------------------------------------------------------------------------</span></a>
|
||||
<a class="sourceLine" id="cb1-2" data-line-number="2"><span class="co"># only_all_tested = FALSE only_all_tested = TRUE</span></a>
|
||||
<a class="sourceLine" id="cb1-3" data-line-number="3"><span class="co"># Antibiotic Antibiotic ----------------------- -----------------------</span></a>
|
||||
<a class="sourceLine" id="cb1-4" data-line-number="4"><span class="co"># A B include as include as include as include as</span></a>
|
||||
<a class="sourceLine" id="cb1-5" data-line-number="5"><span class="co"># numerator denominator numerator denominator</span></a>
|
||||
<a class="sourceLine" id="cb1-6" data-line-number="6"><span class="co"># ---------- ---------- ---------- ----------- ---------- -----------</span></a>
|
||||
<a class="sourceLine" id="cb1-7" data-line-number="7"><span class="co"># S S X X X X</span></a>
|
||||
<a class="sourceLine" id="cb1-8" data-line-number="8"><span class="co"># I S X X X X</span></a>
|
||||
<a class="sourceLine" id="cb1-9" data-line-number="9"><span class="co"># R S X X X X</span></a>
|
||||
<a class="sourceLine" id="cb1-10" data-line-number="10"><span class="co"># not tested S X X - -</span></a>
|
||||
<a class="sourceLine" id="cb1-11" data-line-number="11"><span class="co"># S I X X X X</span></a>
|
||||
<a class="sourceLine" id="cb1-12" data-line-number="12"><span class="co"># I I X X X X</span></a>
|
||||
<a class="sourceLine" id="cb1-13" data-line-number="13"><span class="co"># R I X X X X</span></a>
|
||||
<a class="sourceLine" id="cb1-14" data-line-number="14"><span class="co"># not tested I X X - -</span></a>
|
||||
<a class="sourceLine" id="cb1-15" data-line-number="15"><span class="co"># S R X X X X</span></a>
|
||||
<a class="sourceLine" id="cb1-16" data-line-number="16"><span class="co"># I R X X X X</span></a>
|
||||
<a class="sourceLine" id="cb1-17" data-line-number="17"><span class="co"># R R - X - X</span></a>
|
||||
<a class="sourceLine" id="cb1-18" data-line-number="18"><span class="co"># not tested R - - - -</span></a>
|
||||
<a class="sourceLine" id="cb1-19" data-line-number="19"><span class="co"># S not tested X X - -</span></a>
|
||||
<a class="sourceLine" id="cb1-20" data-line-number="20"><span class="co"># I not tested X X - -</span></a>
|
||||
<a class="sourceLine" id="cb1-21" data-line-number="21"><span class="co"># R not tested - - - -</span></a>
|
||||
<a class="sourceLine" id="cb1-22" data-line-number="22"><span class="co"># not tested not tested - - - -</span></a>
|
||||
<a class="sourceLine" id="cb1-23" data-line-number="23"><span class="co"># -------------------------------------------------------------------------</span></a></code></pre></div>
|
||||
<p>Since this is a major change, usage of the old <code>also_single_tested</code> will throw an informative error that it has been replaced by <code>only_all_tested</code>.</p>
|
||||
</li>
|
||||
</ul>
|
||||
@ -273,23 +273,33 @@
|
||||
<h3 class="hasAnchor">
|
||||
<a href="#changed" class="anchor"></a>Changed</h3>
|
||||
<ul>
|
||||
<li>Fixed a bug in <code><a href="../reference/eucast_rules.html">eucast_rules()</a></code> that caused an error when the input was a specific kind of <code>tibble</code>
|
||||
</li>
|
||||
<li>Removed class <code>atc</code> - using <code><a href="../reference/AMR-deprecated.html">as.atc()</a></code> is now deprecated in favour of <code><a href="../reference/ab_property.html">ab_atc()</a></code> and this will return a character, not the <code>atc</code> class anymore</li>
|
||||
<li>Removed deprecated functions <code>abname()</code>, <code>ab_official()</code>, <code>atc_name()</code>, <code>atc_official()</code>, <code>atc_property()</code>, <code>atc_tradenames()</code>, <code>atc_trivial_nl()</code>
|
||||
</li>
|
||||
<li>Fix and speed improvement for <code><a href="../reference/mo_property.html">mo_shortname()</a></code>
|
||||
</li>
|
||||
<li>Fix for <code><a href="../reference/as.mo.html">as.mo()</a></code> where misspelled input would not be understood</li>
|
||||
<li>Algorithm improvements for <code><a href="../reference/as.mo.html">as.mo()</a></code>:
|
||||
<ul>
|
||||
<li>Some misspelled input were not understood</li>
|
||||
<li>These new trivial names known to the field are now understood: meningococcus, gonococcus, pneumococcus</li>
|
||||
<li>Added support for unknown yeasts and fungi</li>
|
||||
</ul>
|
||||
</li>
|
||||
<li>Added the newest taxonomic data from the IJSEM journal (now up to date until August 2019)</li>
|
||||
<li>Fix for using <code>mo_*</code> functions where the coercion uncertainties and failures would not be available through <code><a href="../reference/as.mo.html">mo_uncertainties()</a></code> and <code><a href="../reference/as.mo.html">mo_failures()</a></code> anymore</li>
|
||||
<li>Deprecated the <code>country</code> parameter of <code><a href="../reference/mdro.html">mdro()</a></code> in favour of the already existing <code>guideline</code> parameter to support multiple guidelines within one country</li>
|
||||
<li>Fix for frequency tables when creating one directly on a group (using <code>group_by()</code>)</li>
|
||||
<li>The name of <code>RIF</code> is now Rifampicin instead of Rifampin</li>
|
||||
<li>The <code>name</code> of <code>RIF</code> is now Rifampicin instead of Rifampin</li>
|
||||
<li>The <code>antibiotics</code> data set is now sorted by name</li>
|
||||
<li>Using verbose mode with <code><a href="../reference/eucast_rules.html">eucast_rules(..., verbose = TRUE)</a></code> returns more informative and readable output</li>
|
||||
<li>Speed improvement for <code><a href="../reference/guess_ab_col.html">guess_ab_col()</a></code> which is now 30 times faster for antibiotic abbreviations</li>
|
||||
</ul>
|
||||
</div>
|
||||
</div>
|
||||
<div id="amr-071" class="section level1">
|
||||
<div id="amr-0-7-1" class="section level1">
|
||||
<h1 class="page-header">
|
||||
<a href="#amr-071" class="anchor"></a>AMR 0.7.1<small> 2019-06-23 </small>
|
||||
<a href="#amr-0-7-1" class="anchor"></a>AMR 0.7.1<small> 2019-06-23 </small>
|
||||
</h1>
|
||||
<div id="new-1" class="section level4">
|
||||
<h4 class="hasAnchor">
|
||||
@ -297,14 +307,14 @@
|
||||
<ul>
|
||||
<li>
|
||||
<p>Function <code><a href="../reference/portion.html">rsi_df()</a></code> to transform a <code>data.frame</code> to a data set containing only the microbial interpretation (S, I, R), the antibiotic, the percentage of S/I/R and the number of available isolates. This is a convenient combination of the existing functions <code><a href="../reference/count.html">count_df()</a></code> and <code><a href="../reference/portion.html">portion_df()</a></code> to immediately show resistance percentages and number of available isolates:</p>
|
||||
<div class="sourceCode" id="cb2"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb2-1" title="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb2-2" title="2"><span class="st"> </span><span class="kw">select</span>(AMX, CIP) <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb2-3" title="3"><span class="st"> </span><span class="kw"><a href="../reference/portion.html">rsi_df</a></span>()</a>
|
||||
<a class="sourceLine" id="cb2-4" title="4"><span class="co"># antibiotic interpretation value isolates</span></a>
|
||||
<a class="sourceLine" id="cb2-5" title="5"><span class="co"># 1 Amoxicillin SI 0.4442636 546</span></a>
|
||||
<a class="sourceLine" id="cb2-6" title="6"><span class="co"># 2 Amoxicillin R 0.5557364 683</span></a>
|
||||
<a class="sourceLine" id="cb2-7" title="7"><span class="co"># 3 Ciprofloxacin SI 0.8381831 1181</span></a>
|
||||
<a class="sourceLine" id="cb2-8" title="8"><span class="co"># 4 Ciprofloxacin R 0.1618169 228</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb2"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb2-1" data-line-number="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb2-2" data-line-number="2"><span class="st"> </span><span class="kw">select</span>(AMX, CIP) <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb2-3" data-line-number="3"><span class="st"> </span><span class="kw"><a href="../reference/portion.html">rsi_df</a></span>()</a>
|
||||
<a class="sourceLine" id="cb2-4" data-line-number="4"><span class="co"># antibiotic interpretation value isolates</span></a>
|
||||
<a class="sourceLine" id="cb2-5" data-line-number="5"><span class="co"># 1 Amoxicillin SI 0.4442636 546</span></a>
|
||||
<a class="sourceLine" id="cb2-6" data-line-number="6"><span class="co"># 2 Amoxicillin R 0.5557364 683</span></a>
|
||||
<a class="sourceLine" id="cb2-7" data-line-number="7"><span class="co"># 3 Ciprofloxacin SI 0.8381831 1181</span></a>
|
||||
<a class="sourceLine" id="cb2-8" data-line-number="8"><span class="co"># 4 Ciprofloxacin R 0.1618169 228</span></a></code></pre></div>
|
||||
</li>
|
||||
<li>
|
||||
<p>Support for all scientifically published pathotypes of <em>E. coli</em> to date (that we could find). Supported are:</p>
|
||||
@ -322,12 +332,12 @@
|
||||
<li>UPEC (Uropathogenic <em>E. coli</em>)</li>
|
||||
</ul>
|
||||
<p>All these lead to the microbial ID of <em>E. coli</em>:</p>
|
||||
<div class="sourceCode" id="cb3"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb3-1" title="1"><span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"UPEC"</span>)</a>
|
||||
<a class="sourceLine" id="cb3-2" title="2"><span class="co"># B_ESCHR_COL</span></a>
|
||||
<a class="sourceLine" id="cb3-3" title="3"><span class="kw"><a href="../reference/mo_property.html">mo_name</a></span>(<span class="st">"UPEC"</span>)</a>
|
||||
<a class="sourceLine" id="cb3-4" title="4"><span class="co"># "Escherichia coli"</span></a>
|
||||
<a class="sourceLine" id="cb3-5" title="5"><span class="kw"><a href="../reference/mo_property.html">mo_gramstain</a></span>(<span class="st">"EHEC"</span>)</a>
|
||||
<a class="sourceLine" id="cb3-6" title="6"><span class="co"># "Gram-negative"</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb3"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb3-1" data-line-number="1"><span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"UPEC"</span>)</a>
|
||||
<a class="sourceLine" id="cb3-2" data-line-number="2"><span class="co"># B_ESCHR_COL</span></a>
|
||||
<a class="sourceLine" id="cb3-3" data-line-number="3"><span class="kw"><a href="../reference/mo_property.html">mo_name</a></span>(<span class="st">"UPEC"</span>)</a>
|
||||
<a class="sourceLine" id="cb3-4" data-line-number="4"><span class="co"># "Escherichia coli"</span></a>
|
||||
<a class="sourceLine" id="cb3-5" data-line-number="5"><span class="kw"><a href="../reference/mo_property.html">mo_gramstain</a></span>(<span class="st">"EHEC"</span>)</a>
|
||||
<a class="sourceLine" id="cb3-6" data-line-number="6"><span class="co"># "Gram-negative"</span></a></code></pre></div>
|
||||
</li>
|
||||
<li>Function <code><a href="../reference/mo_property.html">mo_info()</a></code> as an analogy to <code><a href="../reference/ab_property.html">ab_info()</a></code>. The <code><a href="../reference/mo_property.html">mo_info()</a></code> prints a list with the full taxonomy, authors, and the URL to the online database of a microorganism</li>
|
||||
<li><p>Function <code><a href="../reference/mo_property.html">mo_synonyms()</a></code> to get all previously accepted taxonomic names of a microorganism</p></li>
|
||||
@ -368,9 +378,9 @@
|
||||
</ul>
|
||||
</div>
|
||||
</div>
|
||||
<div id="amr-070" class="section level1">
|
||||
<div id="amr-0-7-0" class="section level1">
|
||||
<h1 class="page-header">
|
||||
<a href="#amr-070" class="anchor"></a>AMR 0.7.0<small> 2019-06-03 </small>
|
||||
<a href="#amr-0-7-0" class="anchor"></a>AMR 0.7.0<small> 2019-06-03 </small>
|
||||
</h1>
|
||||
<div id="new-2" class="section level4">
|
||||
<h4 class="hasAnchor">
|
||||
@ -419,21 +429,21 @@ Please <a href="https://gitlab.com/msberends/AMR/issues/new?issue%5Btitle%5D=Tra
|
||||
<li>The <code><a href="../reference/age.html">age()</a></code> function gained a new parameter <code>exact</code> to determine ages with decimals</li>
|
||||
<li>Removed deprecated functions <code>guess_mo()</code>, <code>guess_atc()</code>, <code>EUCAST_rules()</code>, <code>interpretive_reading()</code>, <code>rsi()</code>
|
||||
</li>
|
||||
<li>Frequency tables (<code><a href="../reference/freq.html">freq()</a></code>):
|
||||
<li>Frequency tables (<code>freq()</code>):
|
||||
<ul>
|
||||
<li>speed improvement for microbial IDs</li>
|
||||
<li>fixed factor level names for R Markdown</li>
|
||||
<li>when all values are unique it now shows a message instead of a warning</li>
|
||||
<li>
|
||||
<p>support for boxplots:</p>
|
||||
<div class="sourceCode" id="cb4"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb4-1" title="1">septic_patients <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb4-2" title="2"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(age) <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb4-3" title="3"><span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/graphics/topics/boxplot">boxplot</a></span>()</a>
|
||||
<a class="sourceLine" id="cb4-4" title="4"><span class="co"># grouped boxplots:</span></a>
|
||||
<a class="sourceLine" id="cb4-5" title="5">septic_patients <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb4-6" title="6"><span class="st"> </span><span class="kw">group_by</span>(hospital_id) <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb4-7" title="7"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(age) <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb4-8" title="8"><span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/graphics/topics/boxplot">boxplot</a></span>()</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb4"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb4-1" data-line-number="1">septic_patients <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb4-2" data-line-number="2"><span class="st"> </span><span class="kw">freq</span>(age) <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb4-3" data-line-number="3"><span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/graphics/topics/boxplot">boxplot</a></span>()</a>
|
||||
<a class="sourceLine" id="cb4-4" data-line-number="4"><span class="co"># grouped boxplots:</span></a>
|
||||
<a class="sourceLine" id="cb4-5" data-line-number="5">septic_patients <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb4-6" data-line-number="6"><span class="st"> </span><span class="kw">group_by</span>(hospital_id) <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb4-7" data-line-number="7"><span class="st"> </span><span class="kw">freq</span>(age) <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb4-8" data-line-number="8"><span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/graphics/topics/boxplot">boxplot</a></span>()</a></code></pre></div>
|
||||
</li>
|
||||
</ul>
|
||||
</li>
|
||||
@ -443,7 +453,7 @@ Please <a href="https://gitlab.com/msberends/AMR/issues/new?issue%5Btitle%5D=EUC
|
||||
<li>Added ceftazidim intrinsic resistance to <em>Streptococci</em>
|
||||
</li>
|
||||
<li>Changed default settings for <code><a href="../reference/age_groups.html">age_groups()</a></code>, to let groups of fives and tens end with 100+ instead of 120+</li>
|
||||
<li>Fix for <code><a href="../reference/freq.html">freq()</a></code> for when all values are <code>NA</code>
|
||||
<li>Fix for <code>freq()</code> for when all values are <code>NA</code>
|
||||
</li>
|
||||
<li>Fix for <code><a href="../reference/first_isolate.html">first_isolate()</a></code> for when dates are missing</li>
|
||||
<li>Improved speed of <code><a href="../reference/guess_ab_col.html">guess_ab_col()</a></code>
|
||||
@ -465,9 +475,9 @@ Please <a href="https://gitlab.com/msberends/AMR/issues/new?issue%5Btitle%5D=EUC
|
||||
</ul>
|
||||
</div>
|
||||
</div>
|
||||
<div id="amr-061" class="section level1">
|
||||
<div id="amr-0-6-1" class="section level1">
|
||||
<h1 class="page-header">
|
||||
<a href="#amr-061" class="anchor"></a>AMR 0.6.1<small> 2019-03-29 </small>
|
||||
<a href="#amr-0-6-1" class="anchor"></a>AMR 0.6.1<small> 2019-03-29 </small>
|
||||
</h1>
|
||||
<div id="changed-3" class="section level4">
|
||||
<h4 class="hasAnchor">
|
||||
@ -479,9 +489,9 @@ Please <a href="https://gitlab.com/msberends/AMR/issues/new?issue%5Btitle%5D=EUC
|
||||
</ul>
|
||||
</div>
|
||||
</div>
|
||||
<div id="amr-060" class="section level1">
|
||||
<div id="amr-0-6-0" class="section level1">
|
||||
<h1 class="page-header">
|
||||
<a href="#amr-060" class="anchor"></a>AMR 0.6.0<small> 2019-03-27 </small>
|
||||
<a href="#amr-0-6-0" class="anchor"></a>AMR 0.6.0<small> 2019-03-27 </small>
|
||||
</h1>
|
||||
<p><strong>New website!</strong></p>
|
||||
<p>We’ve got a new website: <a href="https://msberends.gitlab.io/AMR/">https://msberends.gitlab.io/AMR</a> (built with the great <a href="https://pkgdown.r-lib.org/"><code>pkgdown</code></a>)</p>
|
||||
@ -504,7 +514,7 @@ Please <a href="https://gitlab.com/msberends/AMR/issues/new?issue%5Btitle%5D=EUC
|
||||
<li>
|
||||
<p>The responsible author(s) and year of scientific publication</p>
|
||||
This data is updated annually - check the included version with the new function <code><a href="../reference/catalogue_of_life_version.html">catalogue_of_life_version()</a></code>.</li>
|
||||
<li>Due to this change, some <code>mo</code> codes changed (e.g. <em>Streptococcus</em> changed from <code>B_STRPTC</code> to <code>B_STRPT</code>). A translation table is used internally to support older microorganism IDs, so users will not notice this difference.</li>
|
||||
<li>Due to this change, some <code>mo</code> codes changed (e.g. <em>Streptococcus</em> changed from <code>B_STRPTC</code> to <code>B_STRPT</code>). A translation table is used internally to support older microorganism IDs, so users will not notice this difference.</li>
|
||||
<li>New function <code><a href="../reference/mo_property.html">mo_rank()</a></code> for the taxonomic rank (genus, species, infraspecies, etc.)</li>
|
||||
<li>New function <code><a href="../reference/mo_property.html">mo_url()</a></code> to get the direct URL of a species from the Catalogue of Life</li>
|
||||
</ul>
|
||||
@ -518,33 +528,33 @@ This data is updated annually - check the included version with the new function
|
||||
</li>
|
||||
<li>
|
||||
<p>New filters for antimicrobial classes. Use these functions to filter isolates on results in one of more antibiotics from a specific class:</p>
|
||||
<div class="sourceCode" id="cb5"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb5-1" title="1"><span class="kw"><a href="../reference/filter_ab_class.html">filter_aminoglycosides</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-2" title="2"><span class="kw"><a href="../reference/filter_ab_class.html">filter_carbapenems</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-3" title="3"><span class="kw"><a href="../reference/filter_ab_class.html">filter_cephalosporins</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-4" title="4"><span class="kw"><a href="../reference/filter_ab_class.html">filter_1st_cephalosporins</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-5" title="5"><span class="kw"><a href="../reference/filter_ab_class.html">filter_2nd_cephalosporins</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-6" title="6"><span class="kw"><a href="../reference/filter_ab_class.html">filter_3rd_cephalosporins</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-7" title="7"><span class="kw"><a href="../reference/filter_ab_class.html">filter_4th_cephalosporins</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-8" title="8"><span class="kw"><a href="../reference/filter_ab_class.html">filter_fluoroquinolones</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-9" title="9"><span class="kw"><a href="../reference/filter_ab_class.html">filter_glycopeptides</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-10" title="10"><span class="kw"><a href="../reference/filter_ab_class.html">filter_macrolides</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-11" title="11"><span class="kw"><a href="../reference/filter_ab_class.html">filter_tetracyclines</a></span>()</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb5"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb5-1" data-line-number="1"><span class="kw"><a href="../reference/filter_ab_class.html">filter_aminoglycosides</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-2" data-line-number="2"><span class="kw"><a href="../reference/filter_ab_class.html">filter_carbapenems</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-3" data-line-number="3"><span class="kw"><a href="../reference/filter_ab_class.html">filter_cephalosporins</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-4" data-line-number="4"><span class="kw"><a href="../reference/filter_ab_class.html">filter_1st_cephalosporins</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-5" data-line-number="5"><span class="kw"><a href="../reference/filter_ab_class.html">filter_2nd_cephalosporins</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-6" data-line-number="6"><span class="kw"><a href="../reference/filter_ab_class.html">filter_3rd_cephalosporins</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-7" data-line-number="7"><span class="kw"><a href="../reference/filter_ab_class.html">filter_4th_cephalosporins</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-8" data-line-number="8"><span class="kw"><a href="../reference/filter_ab_class.html">filter_fluoroquinolones</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-9" data-line-number="9"><span class="kw"><a href="../reference/filter_ab_class.html">filter_glycopeptides</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-10" data-line-number="10"><span class="kw"><a href="../reference/filter_ab_class.html">filter_macrolides</a></span>()</a>
|
||||
<a class="sourceLine" id="cb5-11" data-line-number="11"><span class="kw"><a href="../reference/filter_ab_class.html">filter_tetracyclines</a></span>()</a></code></pre></div>
|
||||
<p>The <code>antibiotics</code> data set will be searched, after which the input data will be checked for column names with a value in any abbreviations, codes or official names found in the <code>antibiotics</code> data set. For example:</p>
|
||||
<div class="sourceCode" id="cb6"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb6-1" title="1">septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/filter_ab_class.html">filter_glycopeptides</a></span>(<span class="dt">result =</span> <span class="st">"R"</span>)</a>
|
||||
<a class="sourceLine" id="cb6-2" title="2"><span class="co"># Filtering on glycopeptide antibacterials: any of `vanc` or `teic` is R</span></a>
|
||||
<a class="sourceLine" id="cb6-3" title="3">septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/filter_ab_class.html">filter_glycopeptides</a></span>(<span class="dt">result =</span> <span class="st">"R"</span>, <span class="dt">scope =</span> <span class="st">"all"</span>)</a>
|
||||
<a class="sourceLine" id="cb6-4" title="4"><span class="co"># Filtering on glycopeptide antibacterials: all of `vanc` and `teic` is R</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb6"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb6-1" data-line-number="1">septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/filter_ab_class.html">filter_glycopeptides</a></span>(<span class="dt">result =</span> <span class="st">"R"</span>)</a>
|
||||
<a class="sourceLine" id="cb6-2" data-line-number="2"><span class="co"># Filtering on glycopeptide antibacterials: any of `vanc` or `teic` is R</span></a>
|
||||
<a class="sourceLine" id="cb6-3" data-line-number="3">septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/filter_ab_class.html">filter_glycopeptides</a></span>(<span class="dt">result =</span> <span class="st">"R"</span>, <span class="dt">scope =</span> <span class="st">"all"</span>)</a>
|
||||
<a class="sourceLine" id="cb6-4" data-line-number="4"><span class="co"># Filtering on glycopeptide antibacterials: all of `vanc` and `teic` is R</span></a></code></pre></div>
|
||||
</li>
|
||||
<li>
|
||||
<p>All <code>ab_*</code> functions are deprecated and replaced by <code>atc_*</code> functions:</p>
|
||||
<div class="sourceCode" id="cb7"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb7-1" title="1">ab_property -><span class="st"> </span><span class="kw">atc_property</span>()</a>
|
||||
<a class="sourceLine" id="cb7-2" title="2">ab_name -><span class="st"> </span><span class="kw">atc_name</span>()</a>
|
||||
<a class="sourceLine" id="cb7-3" title="3">ab_official -><span class="st"> </span><span class="kw">atc_official</span>()</a>
|
||||
<a class="sourceLine" id="cb7-4" title="4">ab_trivial_nl -><span class="st"> </span><span class="kw">atc_trivial_nl</span>()</a>
|
||||
<a class="sourceLine" id="cb7-5" title="5">ab_certe -><span class="st"> </span><span class="kw">atc_certe</span>()</a>
|
||||
<a class="sourceLine" id="cb7-6" title="6">ab_umcg -><span class="st"> </span><span class="kw">atc_umcg</span>()</a>
|
||||
<a class="sourceLine" id="cb7-7" title="7">ab_tradenames -><span class="st"> </span><span class="kw">atc_tradenames</span>()</a></code></pre></div>
|
||||
These functions use <code><a href="../reference/AMR-deprecated.html">as.atc()</a></code> internally. The old <code>atc_property</code> has been renamed <code><a href="../reference/atc_online.html">atc_online_property()</a></code>. This is done for two reasons: firstly, not all ATC codes are of antibiotics (ab) but can also be of antivirals or antifungals. Secondly, the input must have class <code>atc</code> or must be coerable to this class. Properties of these classes should start with the same class name, analogous to <code><a href="../reference/as.mo.html">as.mo()</a></code> and e.g. <code>mo_genus</code>.</li>
|
||||
<div class="sourceCode" id="cb7"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb7-1" data-line-number="1">ab_property -><span class="st"> </span><span class="kw">atc_property</span>()</a>
|
||||
<a class="sourceLine" id="cb7-2" data-line-number="2">ab_name -><span class="st"> </span><span class="kw">atc_name</span>()</a>
|
||||
<a class="sourceLine" id="cb7-3" data-line-number="3">ab_official -><span class="st"> </span><span class="kw">atc_official</span>()</a>
|
||||
<a class="sourceLine" id="cb7-4" data-line-number="4">ab_trivial_nl -><span class="st"> </span><span class="kw">atc_trivial_nl</span>()</a>
|
||||
<a class="sourceLine" id="cb7-5" data-line-number="5">ab_certe -><span class="st"> </span><span class="kw">atc_certe</span>()</a>
|
||||
<a class="sourceLine" id="cb7-6" data-line-number="6">ab_umcg -><span class="st"> </span><span class="kw">atc_umcg</span>()</a>
|
||||
<a class="sourceLine" id="cb7-7" data-line-number="7">ab_tradenames -><span class="st"> </span><span class="kw">atc_tradenames</span>()</a></code></pre></div>
|
||||
These functions use <code><a href="../reference/AMR-deprecated.html">as.atc()</a></code> internally. The old <code>atc_property</code> has been renamed <code><a href="../reference/atc_online.html">atc_online_property()</a></code>. This is done for two reasons: firstly, not all ATC codes are of antibiotics (ab) but can also be of antivirals or antifungals. Secondly, the input must have class <code>atc</code> or must be coerable to this class. Properties of these classes should start with the same class name, analogous to <code><a href="../reference/as.mo.html">as.mo()</a></code> and e.g. <code>mo_genus</code>.</li>
|
||||
<li>New functions <code><a href="../reference/mo_source.html">set_mo_source()</a></code> and <code><a href="../reference/mo_source.html">get_mo_source()</a></code> to use your own predefined MO codes as input for <code><a href="../reference/as.mo.html">as.mo()</a></code> and consequently all <code>mo_*</code> functions</li>
|
||||
<li>Support for the upcoming <a href="https://dplyr.tidyverse.org"><code>dplyr</code></a> version 0.8.0</li>
|
||||
<li>New function <code><a href="../reference/guess_ab_col.html">guess_ab_col()</a></code> to find an antibiotic column in a table</li>
|
||||
@ -555,20 +565,20 @@ These functions use <code><a href="../reference/AMR-deprecated.html">as.atc()</a
|
||||
<li>New function <code><a href="../reference/age_groups.html">age_groups()</a></code> to split ages into custom or predefined groups (like children or elderly). This allows for easier demographic antimicrobial resistance analysis per age group.</li>
|
||||
<li>
|
||||
<p>New function <code><a href="../reference/resistance_predict.html">ggplot_rsi_predict()</a></code> as well as the base R <code><a href="https://www.rdocumentation.org/packages/graphics/topics/plot">plot()</a></code> function can now be used for resistance prediction calculated with <code><a href="../reference/resistance_predict.html">resistance_predict()</a></code>:</p>
|
||||
<div class="sourceCode" id="cb8"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb8-1" title="1">x <-<span class="st"> </span><span class="kw"><a href="../reference/resistance_predict.html">resistance_predict</a></span>(septic_patients, <span class="dt">col_ab =</span> <span class="st">"amox"</span>)</a>
|
||||
<a class="sourceLine" id="cb8-2" title="2"><span class="kw"><a href="https://www.rdocumentation.org/packages/graphics/topics/plot">plot</a></span>(x)</a>
|
||||
<a class="sourceLine" id="cb8-3" title="3"><span class="kw"><a href="../reference/resistance_predict.html">ggplot_rsi_predict</a></span>(x)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb8"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb8-1" data-line-number="1">x <-<span class="st"> </span><span class="kw"><a href="../reference/resistance_predict.html">resistance_predict</a></span>(septic_patients, <span class="dt">col_ab =</span> <span class="st">"amox"</span>)</a>
|
||||
<a class="sourceLine" id="cb8-2" data-line-number="2"><span class="kw"><a href="https://www.rdocumentation.org/packages/graphics/topics/plot">plot</a></span>(x)</a>
|
||||
<a class="sourceLine" id="cb8-3" data-line-number="3"><span class="kw"><a href="../reference/resistance_predict.html">ggplot_rsi_predict</a></span>(x)</a></code></pre></div>
|
||||
</li>
|
||||
<li>
|
||||
<p>Functions <code><a href="../reference/first_isolate.html">filter_first_isolate()</a></code> and <code><a href="../reference/first_isolate.html">filter_first_weighted_isolate()</a></code> to shorten and fasten filtering on data sets with antimicrobial results, e.g.:</p>
|
||||
<div class="sourceCode" id="cb9"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb9-1" title="1">septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/first_isolate.html">filter_first_isolate</a></span>(...)</a>
|
||||
<a class="sourceLine" id="cb9-2" title="2"><span class="co"># or</span></a>
|
||||
<a class="sourceLine" id="cb9-3" title="3"><span class="kw"><a href="../reference/first_isolate.html">filter_first_isolate</a></span>(septic_patients, ...)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb9"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb9-1" data-line-number="1">septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/first_isolate.html">filter_first_isolate</a></span>(...)</a>
|
||||
<a class="sourceLine" id="cb9-2" data-line-number="2"><span class="co"># or</span></a>
|
||||
<a class="sourceLine" id="cb9-3" data-line-number="3"><span class="kw"><a href="../reference/first_isolate.html">filter_first_isolate</a></span>(septic_patients, ...)</a></code></pre></div>
|
||||
<p>is equal to:</p>
|
||||
<div class="sourceCode" id="cb10"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb10-1" title="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb10-2" title="2"><span class="st"> </span><span class="kw">mutate</span>(<span class="dt">only_firsts =</span> <span class="kw"><a href="../reference/first_isolate.html">first_isolate</a></span>(septic_patients, ...)) <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb10-3" title="3"><span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/stats/topics/filter">filter</a></span>(only_firsts <span class="op">==</span><span class="st"> </span><span class="ot">TRUE</span>) <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb10-4" title="4"><span class="st"> </span><span class="kw">select</span>(<span class="op">-</span>only_firsts)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb10"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb10-1" data-line-number="1">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb10-2" data-line-number="2"><span class="st"> </span><span class="kw">mutate</span>(<span class="dt">only_firsts =</span> <span class="kw"><a href="../reference/first_isolate.html">first_isolate</a></span>(septic_patients, ...)) <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb10-3" data-line-number="3"><span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/stats/topics/filter">filter</a></span>(only_firsts <span class="op">==</span><span class="st"> </span><span class="ot">TRUE</span>) <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb10-4" data-line-number="4"><span class="st"> </span><span class="kw">select</span>(<span class="op">-</span>only_firsts)</a></code></pre></div>
|
||||
</li>
|
||||
<li>New function <code><a href="../reference/availability.html">availability()</a></code> to check the number of available (non-empty) results in a <code>data.frame</code>
|
||||
</li>
|
||||
@ -597,33 +607,33 @@ These functions use <code><a href="../reference/AMR-deprecated.html">as.atc()</a
|
||||
<ul>
|
||||
<li>
|
||||
<p>Now handles incorrect spelling, like <code>i</code> instead of <code>y</code> and <code>f</code> instead of <code>ph</code>:</p>
|
||||
<div class="sourceCode" id="cb11"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb11-1" title="1"><span class="co"># mo_fullname() uses as.mo() internally</span></a>
|
||||
<a class="sourceLine" id="cb11-2" title="2"></a>
|
||||
<a class="sourceLine" id="cb11-3" title="3"><span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"Sthafilokockus aaureuz"</span>)</a>
|
||||
<a class="sourceLine" id="cb11-4" title="4"><span class="co">#> [1] "Staphylococcus aureus"</span></a>
|
||||
<a class="sourceLine" id="cb11-5" title="5"></a>
|
||||
<a class="sourceLine" id="cb11-6" title="6"><span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"S. klossi"</span>)</a>
|
||||
<a class="sourceLine" id="cb11-7" title="7"><span class="co">#> [1] "Staphylococcus kloosii"</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb11"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb11-1" data-line-number="1"><span class="co"># mo_fullname() uses as.mo() internally</span></a>
|
||||
<a class="sourceLine" id="cb11-2" data-line-number="2"></a>
|
||||
<a class="sourceLine" id="cb11-3" data-line-number="3"><span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"Sthafilokockus aaureuz"</span>)</a>
|
||||
<a class="sourceLine" id="cb11-4" data-line-number="4"><span class="co">#> [1] "Staphylococcus aureus"</span></a>
|
||||
<a class="sourceLine" id="cb11-5" data-line-number="5"></a>
|
||||
<a class="sourceLine" id="cb11-6" data-line-number="6"><span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"S. klossi"</span>)</a>
|
||||
<a class="sourceLine" id="cb11-7" data-line-number="7"><span class="co">#> [1] "Staphylococcus kloosii"</span></a></code></pre></div>
|
||||
</li>
|
||||
<li>
|
||||
<p>Uncertainty of the algorithm is now divided into four levels, 0 to 3, where the default <code>allow_uncertain = TRUE</code> is equal to uncertainty level 2. Run <code><a href="../reference/as.mo.html">?as.mo</a></code> for more info about these levels.</p>
|
||||
<div class="sourceCode" id="cb12"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb12-1" title="1"><span class="co"># equal:</span></a>
|
||||
<a class="sourceLine" id="cb12-2" title="2"><span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(..., <span class="dt">allow_uncertain =</span> <span class="ot">TRUE</span>)</a>
|
||||
<a class="sourceLine" id="cb12-3" title="3"><span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(..., <span class="dt">allow_uncertain =</span> <span class="dv">2</span>)</a>
|
||||
<a class="sourceLine" id="cb12-4" title="4"></a>
|
||||
<a class="sourceLine" id="cb12-5" title="5"><span class="co"># also equal:</span></a>
|
||||
<a class="sourceLine" id="cb12-6" title="6"><span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(..., <span class="dt">allow_uncertain =</span> <span class="ot">FALSE</span>)</a>
|
||||
<a class="sourceLine" id="cb12-7" title="7"><span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(..., <span class="dt">allow_uncertain =</span> <span class="dv">0</span>)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb12"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb12-1" data-line-number="1"><span class="co"># equal:</span></a>
|
||||
<a class="sourceLine" id="cb12-2" data-line-number="2"><span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(..., <span class="dt">allow_uncertain =</span> <span class="ot">TRUE</span>)</a>
|
||||
<a class="sourceLine" id="cb12-3" data-line-number="3"><span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(..., <span class="dt">allow_uncertain =</span> <span class="dv">2</span>)</a>
|
||||
<a class="sourceLine" id="cb12-4" data-line-number="4"></a>
|
||||
<a class="sourceLine" id="cb12-5" data-line-number="5"><span class="co"># also equal:</span></a>
|
||||
<a class="sourceLine" id="cb12-6" data-line-number="6"><span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(..., <span class="dt">allow_uncertain =</span> <span class="ot">FALSE</span>)</a>
|
||||
<a class="sourceLine" id="cb12-7" data-line-number="7"><span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(..., <span class="dt">allow_uncertain =</span> <span class="dv">0</span>)</a></code></pre></div>
|
||||
Using <code><a href="../reference/as.mo.html">as.mo(..., allow_uncertain = 3)</a></code> could lead to very unreliable results.</li>
|
||||
<li>Implemented the latest publication of Becker <em>et al.</em> (2019), for categorising coagulase-negative <em>Staphylococci</em>
|
||||
</li>
|
||||
<li>All microbial IDs that found are now saved to a local file <code>~/.Rhistory_mo</code>. Use the new function <code>clean_mo_history()</code> to delete this file, which resets the algorithms.</li>
|
||||
<li>
|
||||
<p>Incoercible results will now be considered ‘unknown’, MO code <code>UNKNOWN</code>. On foreign systems, properties of these will be translated to all languages already previously supported: German, Dutch, French, Italian, Spanish and Portuguese:</p>
|
||||
<div class="sourceCode" id="cb13"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb13-1" title="1"><span class="kw"><a href="../reference/mo_property.html">mo_genus</a></span>(<span class="st">"qwerty"</span>, <span class="dt">language =</span> <span class="st">"es"</span>)</a>
|
||||
<a class="sourceLine" id="cb13-2" title="2"><span class="co"># Warning: </span></a>
|
||||
<a class="sourceLine" id="cb13-3" title="3"><span class="co"># one unique value (^= 100.0%) could not be coerced and is considered 'unknown': "qwerty". Use mo_failures() to review it.</span></a>
|
||||
<a class="sourceLine" id="cb13-4" title="4"><span class="co">#> [1] "(género desconocido)"</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb13"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb13-1" data-line-number="1"><span class="kw"><a href="../reference/mo_property.html">mo_genus</a></span>(<span class="st">"qwerty"</span>, <span class="dt">language =</span> <span class="st">"es"</span>)</a>
|
||||
<a class="sourceLine" id="cb13-2" data-line-number="2"><span class="co"># Warning: </span></a>
|
||||
<a class="sourceLine" id="cb13-3" data-line-number="3"><span class="co"># one unique value (^= 100.0%) could not be coerced and is considered 'unknown': "qwerty". Use mo_failures() to review it.</span></a>
|
||||
<a class="sourceLine" id="cb13-4" data-line-number="4"><span class="co">#> [1] "(género desconocido)"</span></a></code></pre></div>
|
||||
</li>
|
||||
<li>Fix for vector containing only empty values</li>
|
||||
<li>Finds better results when input is in other languages</li>
|
||||
@ -665,23 +675,23 @@ Using <code><a href="../reference/as.mo.html">as.mo(..., allow_uncertain = 3)</a
|
||||
</li>
|
||||
</ul>
|
||||
</li>
|
||||
<li>Frequency tables (<code><a href="../reference/freq.html">freq()</a></code> function):
|
||||
<li>Frequency tables (<code>freq()</code> function):
|
||||
<ul>
|
||||
<li>
|
||||
<p>Support for tidyverse quasiquotation! Now you can create frequency tables of function outcomes:</p>
|
||||
<div class="sourceCode" id="cb14"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb14-1" title="1"><span class="co"># Determine genus of microorganisms (mo) in `septic_patients` data set:</span></a>
|
||||
<a class="sourceLine" id="cb14-2" title="2"><span class="co"># OLD WAY</span></a>
|
||||
<a class="sourceLine" id="cb14-3" title="3">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb14-4" title="4"><span class="st"> </span><span class="kw">mutate</span>(<span class="dt">genus =</span> <span class="kw"><a href="../reference/mo_property.html">mo_genus</a></span>(mo)) <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb14-5" title="5"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(genus)</a>
|
||||
<a class="sourceLine" id="cb14-6" title="6"><span class="co"># NEW WAY</span></a>
|
||||
<a class="sourceLine" id="cb14-7" title="7">septic_patients <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb14-8" title="8"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(<span class="kw"><a href="../reference/mo_property.html">mo_genus</a></span>(mo))</a>
|
||||
<a class="sourceLine" id="cb14-9" title="9"></a>
|
||||
<a class="sourceLine" id="cb14-10" title="10"><span class="co"># Even supports grouping variables:</span></a>
|
||||
<a class="sourceLine" id="cb14-11" title="11">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb14-12" title="12"><span class="st"> </span><span class="kw">group_by</span>(gender) <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb14-13" title="13"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(<span class="kw"><a href="../reference/mo_property.html">mo_genus</a></span>(mo))</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb14"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb14-1" data-line-number="1"><span class="co"># Determine genus of microorganisms (mo) in `septic_patients` data set:</span></a>
|
||||
<a class="sourceLine" id="cb14-2" data-line-number="2"><span class="co"># OLD WAY</span></a>
|
||||
<a class="sourceLine" id="cb14-3" data-line-number="3">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb14-4" data-line-number="4"><span class="st"> </span><span class="kw">mutate</span>(<span class="dt">genus =</span> <span class="kw"><a href="../reference/mo_property.html">mo_genus</a></span>(mo)) <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb14-5" data-line-number="5"><span class="st"> </span><span class="kw">freq</span>(genus)</a>
|
||||
<a class="sourceLine" id="cb14-6" data-line-number="6"><span class="co"># NEW WAY</span></a>
|
||||
<a class="sourceLine" id="cb14-7" data-line-number="7">septic_patients <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb14-8" data-line-number="8"><span class="st"> </span><span class="kw">freq</span>(<span class="kw"><a href="../reference/mo_property.html">mo_genus</a></span>(mo))</a>
|
||||
<a class="sourceLine" id="cb14-9" data-line-number="9"></a>
|
||||
<a class="sourceLine" id="cb14-10" data-line-number="10"><span class="co"># Even supports grouping variables:</span></a>
|
||||
<a class="sourceLine" id="cb14-11" data-line-number="11">septic_patients <span class="op">%>%</span></a>
|
||||
<a class="sourceLine" id="cb14-12" data-line-number="12"><span class="st"> </span><span class="kw">group_by</span>(gender) <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb14-13" data-line-number="13"><span class="st"> </span><span class="kw">freq</span>(<span class="kw"><a href="../reference/mo_property.html">mo_genus</a></span>(mo))</a></code></pre></div>
|
||||
</li>
|
||||
<li>Header info is now available as a list, with the <code>header</code> function</li>
|
||||
<li>The parameter <code>header</code> is now set to <code>TRUE</code> at default, even for markdown</li>
|
||||
@ -712,9 +722,9 @@ Using <code><a href="../reference/as.mo.html">as.mo(..., allow_uncertain = 3)</a
|
||||
</ul>
|
||||
</div>
|
||||
</div>
|
||||
<div id="amr-050" class="section level1">
|
||||
<div id="amr-0-5-0" class="section level1">
|
||||
<h1 class="page-header">
|
||||
<a href="#amr-050" class="anchor"></a>AMR 0.5.0<small> 2018-11-30 </small>
|
||||
<a href="#amr-0-5-0" class="anchor"></a>AMR 0.5.0<small> 2018-11-30 </small>
|
||||
</h1>
|
||||
<div id="new-4" class="section level4">
|
||||
<h4 class="hasAnchor">
|
||||
@ -756,10 +766,10 @@ Using <code><a href="../reference/as.mo.html">as.mo(..., allow_uncertain = 3)</a
|
||||
<li>Fewer than 3 characters as input for <code>as.mo</code> will return NA</li>
|
||||
<li>
|
||||
<p>Function <code>as.mo</code> (and all <code>mo_*</code> wrappers) now supports genus abbreviations with “species” attached</p>
|
||||
<div class="sourceCode" id="cb15"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb15-1" title="1"><span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"E. species"</span>) <span class="co"># B_ESCHR</span></a>
|
||||
<a class="sourceLine" id="cb15-2" title="2"><span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"E. spp."</span>) <span class="co"># "Escherichia species"</span></a>
|
||||
<a class="sourceLine" id="cb15-3" title="3"><span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"S. spp"</span>) <span class="co"># B_STPHY</span></a>
|
||||
<a class="sourceLine" id="cb15-4" title="4"><span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"S. species"</span>) <span class="co"># "Staphylococcus species"</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb15"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb15-1" data-line-number="1"><span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"E. species"</span>) <span class="co"># B_ESCHR</span></a>
|
||||
<a class="sourceLine" id="cb15-2" data-line-number="2"><span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"E. spp."</span>) <span class="co"># "Escherichia species"</span></a>
|
||||
<a class="sourceLine" id="cb15-3" data-line-number="3"><span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"S. spp"</span>) <span class="co"># B_STPHY</span></a>
|
||||
<a class="sourceLine" id="cb15-4" data-line-number="4"><span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"S. species"</span>) <span class="co"># "Staphylococcus species"</span></a></code></pre></div>
|
||||
</li>
|
||||
<li>Added parameter <code>combine_IR</code> (TRUE/FALSE) to functions <code>portion_df</code> and <code>count_df</code>, to indicate that all values of I and R must be merged into one, so the output only consists of S vs. IR (susceptible vs. non-susceptible)</li>
|
||||
<li>Fix for <code>portion_*(..., as_percent = TRUE)</code> when minimal number of isolates would not be met</li>
|
||||
@ -768,21 +778,21 @@ Using <code><a href="../reference/as.mo.html">as.mo(..., allow_uncertain = 3)</a
|
||||
<li>Using <code>portion_*</code> functions now throws a warning when total available isolate is below parameter <code>minimum</code>
|
||||
</li>
|
||||
<li>Functions <code>as.mo</code>, <code>as.rsi</code>, <code>as.mic</code>, <code>as.atc</code> and <code>freq</code> will not set package name as attribute anymore</li>
|
||||
<li>Frequency tables - <code><a href="../reference/freq.html">freq()</a></code>:
|
||||
<li>Frequency tables - <code>freq()</code>:
|
||||
<ul>
|
||||
<li>
|
||||
<p>Support for grouping variables, test with:</p>
|
||||
<div class="sourceCode" id="cb16"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb16-1" title="1">septic_patients <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb16-2" title="2"><span class="st"> </span><span class="kw">group_by</span>(hospital_id) <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb16-3" title="3"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(gender)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb16"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb16-1" data-line-number="1">septic_patients <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb16-2" data-line-number="2"><span class="st"> </span><span class="kw">group_by</span>(hospital_id) <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb16-3" data-line-number="3"><span class="st"> </span><span class="kw">freq</span>(gender)</a></code></pre></div>
|
||||
</li>
|
||||
<li>
|
||||
<p>Support for (un)selecting columns:</p>
|
||||
<div class="sourceCode" id="cb17"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb17-1" title="1">septic_patients <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb17-2" title="2"><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(hospital_id) <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb17-3" title="3"><span class="st"> </span><span class="kw">select</span>(<span class="op">-</span>count, <span class="op">-</span>cum_count) <span class="co"># only get item, percent, cum_percent</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb17"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb17-1" data-line-number="1">septic_patients <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb17-2" data-line-number="2"><span class="st"> </span><span class="kw">freq</span>(hospital_id) <span class="op">%>%</span><span class="st"> </span></a>
|
||||
<a class="sourceLine" id="cb17-3" data-line-number="3"><span class="st"> </span><span class="kw">select</span>(<span class="op">-</span>count, <span class="op">-</span>cum_count) <span class="co"># only get item, percent, cum_percent</span></a></code></pre></div>
|
||||
</li>
|
||||
<li>Check for <code><a href="https://www.rdocumentation.org/packages/hms/topics/hms">hms::is.hms</a></code>
|
||||
<li>Check for <code><a href="https://www.rdocumentation.org/packages/hms/topics/Deprecated">hms::is.hms</a></code>
|
||||
</li>
|
||||
<li>Now prints in markdown at default in non-interactive sessions</li>
|
||||
<li>No longer adds the factor level column and sorts factors on count again</li>
|
||||
@ -800,7 +810,7 @@ Using <code><a href="../reference/as.mo.html">as.mo(..., allow_uncertain = 3)</a
|
||||
<li>Removed diacritics from all authors (columns <code>microorganisms$ref</code> and <code>microorganisms.old$ref</code>) to comply with CRAN policy to only allow ASCII characters</li>
|
||||
<li>Fix for <code>mo_property</code> not working properly</li>
|
||||
<li>Fix for <code>eucast_rules</code> where some Streptococci would become ceftazidime R in EUCAST rule 4.5</li>
|
||||
<li>Support for named vectors of class <code>mo</code>, useful for <code><a href="../reference/freq.html">top_freq()</a></code>
|
||||
<li>Support for named vectors of class <code>mo</code>, useful for <code>top_freq()</code>
|
||||
</li>
|
||||
<li>
|
||||
<code>ggplot_rsi</code> and <code>scale_y_percent</code> have <code>breaks</code> parameter</li>
|
||||
@ -839,9 +849,9 @@ Using <code><a href="../reference/as.mo.html">as.mo(..., allow_uncertain = 3)</a
|
||||
</ul>
|
||||
</div>
|
||||
</div>
|
||||
<div id="amr-040" class="section level1">
|
||||
<div id="amr-0-4-0" class="section level1">
|
||||
<h1 class="page-header">
|
||||
<a href="#amr-040" class="anchor"></a>AMR 0.4.0<small> 2018-10-01 </small>
|
||||
<a href="#amr-0-4-0" class="anchor"></a>AMR 0.4.0<small> 2018-10-01 </small>
|
||||
</h1>
|
||||
<div id="new-5" class="section level4">
|
||||
<h4 class="hasAnchor">
|
||||
@ -860,18 +870,18 @@ Using <code><a href="../reference/as.mo.html">as.mo(..., allow_uncertain = 3)</a
|
||||
</li>
|
||||
</ul>
|
||||
<p>They also come with support for German, Dutch, French, Italian, Spanish and Portuguese:</p>
|
||||
<div class="sourceCode" id="cb18"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb18-1" title="1"><span class="kw"><a href="../reference/mo_property.html">mo_gramstain</a></span>(<span class="st">"E. coli"</span>)</a>
|
||||
<a class="sourceLine" id="cb18-2" title="2"><span class="co"># [1] "Gram negative"</span></a>
|
||||
<a class="sourceLine" id="cb18-3" title="3"><span class="kw"><a href="../reference/mo_property.html">mo_gramstain</a></span>(<span class="st">"E. coli"</span>, <span class="dt">language =</span> <span class="st">"de"</span>) <span class="co"># German</span></a>
|
||||
<a class="sourceLine" id="cb18-4" title="4"><span class="co"># [1] "Gramnegativ"</span></a>
|
||||
<a class="sourceLine" id="cb18-5" title="5"><span class="kw"><a href="../reference/mo_property.html">mo_gramstain</a></span>(<span class="st">"E. coli"</span>, <span class="dt">language =</span> <span class="st">"es"</span>) <span class="co"># Spanish</span></a>
|
||||
<a class="sourceLine" id="cb18-6" title="6"><span class="co"># [1] "Gram negativo"</span></a>
|
||||
<a class="sourceLine" id="cb18-7" title="7"><span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"S. group A"</span>, <span class="dt">language =</span> <span class="st">"pt"</span>) <span class="co"># Portuguese</span></a>
|
||||
<a class="sourceLine" id="cb18-8" title="8"><span class="co"># [1] "Streptococcus grupo A"</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb18"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb18-1" data-line-number="1"><span class="kw"><a href="../reference/mo_property.html">mo_gramstain</a></span>(<span class="st">"E. coli"</span>)</a>
|
||||
<a class="sourceLine" id="cb18-2" data-line-number="2"><span class="co"># [1] "Gram negative"</span></a>
|
||||
<a class="sourceLine" id="cb18-3" data-line-number="3"><span class="kw"><a href="../reference/mo_property.html">mo_gramstain</a></span>(<span class="st">"E. coli"</span>, <span class="dt">language =</span> <span class="st">"de"</span>) <span class="co"># German</span></a>
|
||||
<a class="sourceLine" id="cb18-4" data-line-number="4"><span class="co"># [1] "Gramnegativ"</span></a>
|
||||
<a class="sourceLine" id="cb18-5" data-line-number="5"><span class="kw"><a href="../reference/mo_property.html">mo_gramstain</a></span>(<span class="st">"E. coli"</span>, <span class="dt">language =</span> <span class="st">"es"</span>) <span class="co"># Spanish</span></a>
|
||||
<a class="sourceLine" id="cb18-6" data-line-number="6"><span class="co"># [1] "Gram negativo"</span></a>
|
||||
<a class="sourceLine" id="cb18-7" data-line-number="7"><span class="kw"><a href="../reference/mo_property.html">mo_fullname</a></span>(<span class="st">"S. group A"</span>, <span class="dt">language =</span> <span class="st">"pt"</span>) <span class="co"># Portuguese</span></a>
|
||||
<a class="sourceLine" id="cb18-8" data-line-number="8"><span class="co"># [1] "Streptococcus grupo A"</span></a></code></pre></div>
|
||||
<p>Furthermore, former taxonomic names will give a note about the current taxonomic name:</p>
|
||||
<div class="sourceCode" id="cb19"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb19-1" title="1"><span class="kw"><a href="../reference/mo_property.html">mo_gramstain</a></span>(<span class="st">"Esc blattae"</span>)</a>
|
||||
<a class="sourceLine" id="cb19-2" title="2"><span class="co"># Note: 'Escherichia blattae' (Burgess et al., 1973) was renamed 'Shimwellia blattae' (Priest and Barker, 2010)</span></a>
|
||||
<a class="sourceLine" id="cb19-3" title="3"><span class="co"># [1] "Gram negative"</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb19"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb19-1" data-line-number="1"><span class="kw"><a href="../reference/mo_property.html">mo_gramstain</a></span>(<span class="st">"Esc blattae"</span>)</a>
|
||||
<a class="sourceLine" id="cb19-2" data-line-number="2"><span class="co"># Note: 'Escherichia blattae' (Burgess et al., 1973) was renamed 'Shimwellia blattae' (Priest and Barker, 2010)</span></a>
|
||||
<a class="sourceLine" id="cb19-3" data-line-number="3"><span class="co"># [1] "Gram negative"</span></a></code></pre></div>
|
||||
</li>
|
||||
<li>Functions <code>count_R</code>, <code>count_IR</code>, <code>count_I</code>, <code>count_SI</code> and <code>count_S</code> to selectively count resistant or susceptible isolates
|
||||
<ul>
|
||||
@ -882,18 +892,18 @@ Using <code><a href="../reference/as.mo.html">as.mo(..., allow_uncertain = 3)</a
|
||||
</li>
|
||||
<li>
|
||||
<p>Functions <code>as.mo</code> and <code>is.mo</code> as replacements for <code>as.bactid</code> and <code>is.bactid</code> (since the <code>microoganisms</code> data set not only contains bacteria). These last two functions are deprecated and will be removed in a future release. The <code>as.mo</code> function determines microbial IDs using intelligent rules:</p>
|
||||
<div class="sourceCode" id="cb20"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb20-1" title="1"><span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"E. coli"</span>)</a>
|
||||
<a class="sourceLine" id="cb20-2" title="2"><span class="co"># [1] B_ESCHR_COL</span></a>
|
||||
<a class="sourceLine" id="cb20-3" title="3"><span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"MRSA"</span>)</a>
|
||||
<a class="sourceLine" id="cb20-4" title="4"><span class="co"># [1] B_STPHY_AUR</span></a>
|
||||
<a class="sourceLine" id="cb20-5" title="5"><span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"S group A"</span>)</a>
|
||||
<a class="sourceLine" id="cb20-6" title="6"><span class="co"># [1] B_STRPTC_GRA</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb20"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb20-1" data-line-number="1"><span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"E. coli"</span>)</a>
|
||||
<a class="sourceLine" id="cb20-2" data-line-number="2"><span class="co"># [1] B_ESCHR_COL</span></a>
|
||||
<a class="sourceLine" id="cb20-3" data-line-number="3"><span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"MRSA"</span>)</a>
|
||||
<a class="sourceLine" id="cb20-4" data-line-number="4"><span class="co"># [1] B_STPHY_AUR</span></a>
|
||||
<a class="sourceLine" id="cb20-5" data-line-number="5"><span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(<span class="st">"S group A"</span>)</a>
|
||||
<a class="sourceLine" id="cb20-6" data-line-number="6"><span class="co"># [1] B_STRPTC_GRA</span></a></code></pre></div>
|
||||
<p>And with great speed too - on a quite regular Linux server from 2007 it takes us less than 0.02 seconds to transform 25,000 items:</p>
|
||||
<div class="sourceCode" id="cb21"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb21-1" title="1">thousands_of_E_colis <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/rep">rep</a></span>(<span class="st">"E. coli"</span>, <span class="dv">25000</span>)</a>
|
||||
<a class="sourceLine" id="cb21-2" title="2">microbenchmark<span class="op">::</span><span class="kw"><a href="https://www.rdocumentation.org/packages/microbenchmark/topics/microbenchmark">microbenchmark</a></span>(<span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(thousands_of_E_colis), <span class="dt">unit =</span> <span class="st">"s"</span>)</a>
|
||||
<a class="sourceLine" id="cb21-3" title="3"><span class="co"># Unit: seconds</span></a>
|
||||
<a class="sourceLine" id="cb21-4" title="4"><span class="co"># min median max neval</span></a>
|
||||
<a class="sourceLine" id="cb21-5" title="5"><span class="co"># 0.01817717 0.01843957 0.03878077 100</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb21"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb21-1" data-line-number="1">thousands_of_E_colis <-<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/rep">rep</a></span>(<span class="st">"E. coli"</span>, <span class="dv">25000</span>)</a>
|
||||
<a class="sourceLine" id="cb21-2" data-line-number="2">microbenchmark<span class="op">::</span><span class="kw"><a href="https://www.rdocumentation.org/packages/microbenchmark/topics/microbenchmark">microbenchmark</a></span>(<span class="kw"><a href="../reference/as.mo.html">as.mo</a></span>(thousands_of_E_colis), <span class="dt">unit =</span> <span class="st">"s"</span>)</a>
|
||||
<a class="sourceLine" id="cb21-3" data-line-number="3"><span class="co"># Unit: seconds</span></a>
|
||||
<a class="sourceLine" id="cb21-4" data-line-number="4"><span class="co"># min median max neval</span></a>
|
||||
<a class="sourceLine" id="cb21-5" data-line-number="5"><span class="co"># 0.01817717 0.01843957 0.03878077 100</span></a></code></pre></div>
|
||||
</li>
|
||||
<li>Added parameter <code>reference_df</code> for <code>as.mo</code>, so users can supply their own microbial IDs, name or codes as a reference table</li>
|
||||
<li>Renamed all previous references to <code>bactid</code> to <code>mo</code>, like:
|
||||
@ -921,12 +931,12 @@ Using <code><a href="../reference/as.mo.html">as.mo(..., allow_uncertain = 3)</a
|
||||
<li>Added three antimicrobial agents to the <code>antibiotics</code> data set: Terbinafine (D01BA02), Rifaximin (A07AA11) and Isoconazole (D01AC05)</li>
|
||||
<li>
|
||||
<p>Added 163 trade names to the <code>antibiotics</code> data set, it now contains 298 different trade names in total, e.g.:</p>
|
||||
<div class="sourceCode" id="cb22"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb22-1" title="1"><span class="kw">ab_official</span>(<span class="st">"Bactroban"</span>)</a>
|
||||
<a class="sourceLine" id="cb22-2" title="2"><span class="co"># [1] "Mupirocin"</span></a>
|
||||
<a class="sourceLine" id="cb22-3" title="3"><span class="kw"><a href="../reference/ab_property.html">ab_name</a></span>(<span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/c">c</a></span>(<span class="st">"Bactroban"</span>, <span class="st">"Amoxil"</span>, <span class="st">"Zithromax"</span>, <span class="st">"Floxapen"</span>))</a>
|
||||
<a class="sourceLine" id="cb22-4" title="4"><span class="co"># [1] "Mupirocin" "Amoxicillin" "Azithromycin" "Flucloxacillin"</span></a>
|
||||
<a class="sourceLine" id="cb22-5" title="5"><span class="kw"><a href="../reference/ab_property.html">ab_atc</a></span>(<span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/c">c</a></span>(<span class="st">"Bactroban"</span>, <span class="st">"Amoxil"</span>, <span class="st">"Zithromax"</span>, <span class="st">"Floxapen"</span>))</a>
|
||||
<a class="sourceLine" id="cb22-6" title="6"><span class="co"># [1] "R01AX06" "J01CA04" "J01FA10" "J01CF05"</span></a></code></pre></div>
|
||||
<div class="sourceCode" id="cb22"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb22-1" data-line-number="1"><span class="kw">ab_official</span>(<span class="st">"Bactroban"</span>)</a>
|
||||
<a class="sourceLine" id="cb22-2" data-line-number="2"><span class="co"># [1] "Mupirocin"</span></a>
|
||||
<a class="sourceLine" id="cb22-3" data-line-number="3"><span class="kw"><a href="../reference/ab_property.html">ab_name</a></span>(<span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/c">c</a></span>(<span class="st">"Bactroban"</span>, <span class="st">"Amoxil"</span>, <span class="st">"Zithromax"</span>, <span class="st">"Floxapen"</span>))</a>
|
||||
<a class="sourceLine" id="cb22-4" data-line-number="4"><span class="co"># [1] "Mupirocin" "Amoxicillin" "Azithromycin" "Flucloxacillin"</span></a>
|
||||
<a class="sourceLine" id="cb22-5" data-line-number="5"><span class="kw"><a href="../reference/ab_property.html">ab_atc</a></span>(<span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/c">c</a></span>(<span class="st">"Bactroban"</span>, <span class="st">"Amoxil"</span>, <span class="st">"Zithromax"</span>, <span class="st">"Floxapen"</span>))</a>
|
||||
<a class="sourceLine" id="cb22-6" data-line-number="6"><span class="co"># [1] "R01AX06" "J01CA04" "J01FA10" "J01CF05"</span></a></code></pre></div>
|
||||
</li>
|
||||
<li>For <code>first_isolate</code>, rows will be ignored when there’s no species available</li>
|
||||
<li>Function <code>ratio</code> is now deprecated and will be removed in a future release, as it is not really the scope of this package</li>
|
||||
@ -937,13 +947,13 @@ Using <code><a href="../reference/as.mo.html">as.mo(..., allow_uncertain = 3)</a
|
||||
</li>
|
||||
<li>
|
||||
<p>Support for quasiquotation in the functions series <code>count_*</code> and <code>portions_*</code>, and <code>n_rsi</code>. This allows to check for more than 2 vectors or columns.</p>
|
||||
<div class="sourceCode" id="cb23"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb23-1" title="1">septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw">select</span>(amox, cipr) <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/count.html">count_IR</a></span>()</a>
|
||||
<a class="sourceLine" id="cb23-2" title="2"><span class="co"># which is the same as:</span></a>
|
||||
<a class="sourceLine" id="cb23-3" title="3">septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/count.html">count_IR</a></span>(amox, cipr)</a>
|
||||
<a class="sourceLine" id="cb23-4" title="4"></a>
|
||||
<a class="sourceLine" id="cb23-5" title="5">septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/portion.html">portion_S</a></span>(amcl)</a>
|
||||
<a class="sourceLine" id="cb23-6" title="6">septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/portion.html">portion_S</a></span>(amcl, gent)</a>
|
||||
<a class="sourceLine" id="cb23-7" title="7">septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/portion.html">portion_S</a></span>(amcl, gent, pita)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb23"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb23-1" data-line-number="1">septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw">select</span>(amox, cipr) <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/count.html">count_IR</a></span>()</a>
|
||||
<a class="sourceLine" id="cb23-2" data-line-number="2"><span class="co"># which is the same as:</span></a>
|
||||
<a class="sourceLine" id="cb23-3" data-line-number="3">septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/count.html">count_IR</a></span>(amox, cipr)</a>
|
||||
<a class="sourceLine" id="cb23-4" data-line-number="4"></a>
|
||||
<a class="sourceLine" id="cb23-5" data-line-number="5">septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/portion.html">portion_S</a></span>(amcl)</a>
|
||||
<a class="sourceLine" id="cb23-6" data-line-number="6">septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/portion.html">portion_S</a></span>(amcl, gent)</a>
|
||||
<a class="sourceLine" id="cb23-7" data-line-number="7">septic_patients <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/portion.html">portion_S</a></span>(amcl, gent, pita)</a></code></pre></div>
|
||||
</li>
|
||||
<li>Edited <code>ggplot_rsi</code> and <code>geom_rsi</code> so they can cope with <code>count_df</code>. The new <code>fun</code> parameter has value <code>portion_df</code> at default, but can be set to <code>count_df</code>.</li>
|
||||
<li>Fix for <code>ggplot_rsi</code> when the <code>ggplot2</code> package was not loaded</li>
|
||||
@ -957,12 +967,12 @@ Using <code><a href="../reference/as.mo.html">as.mo(..., allow_uncertain = 3)</a
|
||||
</li>
|
||||
<li>
|
||||
<p>Support for types (classes) list and matrix for <code>freq</code></p>
|
||||
<div class="sourceCode" id="cb24"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb24-1" title="1">my_matrix =<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/with">with</a></span>(septic_patients, <span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/matrix">matrix</a></span>(<span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/c">c</a></span>(age, gender), <span class="dt">ncol =</span> <span class="dv">2</span>))</a>
|
||||
<a class="sourceLine" id="cb24-2" title="2"><span class="kw"><a href="../reference/freq.html">freq</a></span>(my_matrix)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb24"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb24-1" data-line-number="1">my_matrix =<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/with">with</a></span>(septic_patients, <span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/matrix">matrix</a></span>(<span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/c">c</a></span>(age, gender), <span class="dt">ncol =</span> <span class="dv">2</span>))</a>
|
||||
<a class="sourceLine" id="cb24-2" data-line-number="2"><span class="kw">freq</span>(my_matrix)</a></code></pre></div>
|
||||
<p>For lists, subsetting is possible:</p>
|
||||
<div class="sourceCode" id="cb25"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb25-1" title="1">my_list =<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/list">list</a></span>(<span class="dt">age =</span> septic_patients<span class="op">$</span>age, <span class="dt">gender =</span> septic_patients<span class="op">$</span>gender)</a>
|
||||
<a class="sourceLine" id="cb25-2" title="2">my_list <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(age)</a>
|
||||
<a class="sourceLine" id="cb25-3" title="3">my_list <span class="op">%>%</span><span class="st"> </span><span class="kw"><a href="../reference/freq.html">freq</a></span>(gender)</a></code></pre></div>
|
||||
<div class="sourceCode" id="cb25"><pre class="sourceCode r"><code class="sourceCode r"><a class="sourceLine" id="cb25-1" data-line-number="1">my_list =<span class="st"> </span><span class="kw"><a href="https://www.rdocumentation.org/packages/base/topics/list">list</a></span>(<span class="dt">age =</span> septic_patients<span class="op">$</span>age, <span class="dt">gender =</span> septic_patients<span class="op">$</span>gender)</a>
|
||||
<a class="sourceLine" id="cb25-2" data-line-number="2">my_list <span class="op">%>%</span><span class="st"> </span><span class="kw">freq</span>(age)</a>
|
||||
<a class="sourceLine" id="cb25-3" data-line-number="3">my_list <span class="op">%>%</span><span class="st"> </span><span class="kw">freq</span>(gender)</a></code></pre></div>
|
||||
</li>
|
||||
</ul>
|
||||
</div>
|
||||
@ -974,9 +984,9 @@ Using <code><a href="../reference/as.mo.html">as.mo(..., allow_uncertain = 3)</a
|
||||
</ul>
|
||||
</div>
|
||||
</div>
|
||||
<div id="amr-030" class="section level1">
|
||||
<div id="amr-0-3-0" class="section level1">
|
||||
<h1 class="page-header">
|
||||
<a href="#amr-030" class="anchor"></a>AMR 0.3.0<small> 2018-08-14 </small>
|
||||
<a href="#amr-0-3-0" class="anchor"></a>AMR 0.3.0<small> 2018-08-14 </small>
|
||||
</h1>
|
||||
<div id="new-6" class="section level4">
|
||||
<h4 class="hasAnchor">
|
||||
@ -1036,13 +1046,13 @@ Using <code><a href="../reference/as.mo.html">as.mo(..., allow_uncertain = 3)</a
|
||||
<ul>
|
||||
<li>A vignette to explain its usage</li>
|
||||
<li>Support for <code>rsi</code> (antimicrobial resistance) to use as input</li>
|
||||
<li>Support for <code>table</code> to use as input: <code><a href="../reference/freq.html">freq(table(x, y))</a></code>
|
||||
<li>Support for <code>table</code> to use as input: <code>freq(table(x, y))</code>
|
||||
</li>
|
||||
<li>Support for existing functions <code>hist</code> and <code>plot</code> to use a frequency table as input: <code><a href="https://www.rdocumentation.org/packages/graphics/topics/hist">hist(freq(df$age))</a></code>
|
||||
</li>
|
||||
<li>Support for <code>as.vector</code>, <code>as.data.frame</code>, <code>as_tibble</code> and <code>format</code>
|
||||
</li>
|
||||
<li>Support for quasiquotation: <code><a href="../reference/freq.html">freq(mydata, mycolumn)</a></code> is the same as <code>mydata %>% freq(mycolumn)</code>
|
||||
<li>Support for quasiquotation: <code>freq(mydata, mycolumn)</code> is the same as <code>mydata %>% freq(mycolumn)</code>
|
||||
</li>
|
||||
<li>Function <code>top_freq</code> function to return the top/below <em>n</em> items as vector</li>
|
||||
<li>Header of frequency tables now also show Mean Absolute Deviaton (MAD) and Interquartile Range (IQR)</li>
|
||||
@ -1081,7 +1091,7 @@ Using <code><a href="../reference/as.mo.html">as.mo(..., allow_uncertain = 3)</a
|
||||
</li>
|
||||
</ul>
|
||||
</li>
|
||||
<li>Now possible to coerce MIC values with a space between operator and value, i.e. <code><a href="../reference/as.mic.html">as.mic("<= 0.002")</a></code> now works</li>
|
||||
<li>Now possible to coerce MIC values with a space between operator and value, i.e. <code><a href="../reference/as.mic.html">as.mic("<= 0.002")</a></code> now works</li>
|
||||
<li>Classes <code>rsi</code> and <code>mic</code> do not add the attribute <code>package.version</code> anymore</li>
|
||||
<li>Added <code>"groups"</code> option for <code>atc_property(..., property)</code>. It will return a vector of the ATC hierarchy as defined by the <a href="https://www.whocc.no/atc/structure_and_principles/">WHO</a>. The new function <code>atc_groups</code> is a convenient wrapper around this.</li>
|
||||
<li>Build-in host check for <code>atc_property</code> as it requires the host set by <code>url</code> to be responsive</li>
|
||||
@ -1111,9 +1121,9 @@ Using <code><a href="../reference/as.mo.html">as.mo(..., allow_uncertain = 3)</a
|
||||
</ul>
|
||||
</div>
|
||||
</div>
|
||||
<div id="amr-020" class="section level1">
|
||||
<div id="amr-0-2-0" class="section level1">
|
||||
<h1 class="page-header">
|
||||
<a href="#amr-020" class="anchor"></a>AMR 0.2.0<small> 2018-05-03 </small>
|
||||
<a href="#amr-0-2-0" class="anchor"></a>AMR 0.2.0<small> 2018-05-03 </small>
|
||||
</h1>
|
||||
<div id="new-7" class="section level4">
|
||||
<h4 class="hasAnchor">
|
||||
@ -1169,9 +1179,9 @@ Using <code><a href="../reference/as.mo.html">as.mo(..., allow_uncertain = 3)</a
|
||||
</ul>
|
||||
</div>
|
||||
</div>
|
||||
<div id="amr-011" class="section level1">
|
||||
<div id="amr-0-1-1" class="section level1">
|
||||
<h1 class="page-header">
|
||||
<a href="#amr-011" class="anchor"></a>AMR 0.1.1<small> 2018-03-14 </small>
|
||||
<a href="#amr-0-1-1" class="anchor"></a>AMR 0.1.1<small> 2018-03-14 </small>
|
||||
</h1>
|
||||
<ul>
|
||||
<li>
|
||||
@ -1182,9 +1192,9 @@ Using <code><a href="../reference/as.mo.html">as.mo(..., allow_uncertain = 3)</a
|
||||
<li>Added barplots for <code>rsi</code> and <code>mic</code> classes</li>
|
||||
</ul>
|
||||
</div>
|
||||
<div id="amr-010" class="section level1">
|
||||
<div id="amr-0-1-0" class="section level1">
|
||||
<h1 class="page-header">
|
||||
<a href="#amr-010" class="anchor"></a>AMR 0.1.0<small> 2018-02-22 </small>
|
||||
<a href="#amr-0-1-0" class="anchor"></a>AMR 0.1.0<small> 2018-02-22 </small>
|
||||
</h1>
|
||||
<ul>
|
||||
<li>First submission to CRAN.</li>
|
||||
@ -1196,17 +1206,17 @@ Using <code><a href="../reference/as.mo.html">as.mo(..., allow_uncertain = 3)</a
|
||||
<div id="tocnav">
|
||||
<h2>Contents</h2>
|
||||
<ul class="nav nav-pills nav-stacked">
|
||||
<li><a href="#amr-0719008">0.7.1.9008</a></li>
|
||||
<li><a href="#amr-071">0.7.1</a></li>
|
||||
<li><a href="#amr-070">0.7.0</a></li>
|
||||
<li><a href="#amr-061">0.6.1</a></li>
|
||||
<li><a href="#amr-060">0.6.0</a></li>
|
||||
<li><a href="#amr-050">0.5.0</a></li>
|
||||
<li><a href="#amr-040">0.4.0</a></li>
|
||||
<li><a href="#amr-030">0.3.0</a></li>
|
||||
<li><a href="#amr-020">0.2.0</a></li>
|
||||
<li><a href="#amr-011">0.1.1</a></li>
|
||||
<li><a href="#amr-010">0.1.0</a></li>
|
||||
<li><a href="#amr-0-7-1-9026">0.7.1.9026</a></li>
|
||||
<li><a href="#amr-0-7-1">0.7.1</a></li>
|
||||
<li><a href="#amr-0-7-0">0.7.0</a></li>
|
||||
<li><a href="#amr-0-6-1">0.6.1</a></li>
|
||||
<li><a href="#amr-0-6-0">0.6.0</a></li>
|
||||
<li><a href="#amr-0-5-0">0.5.0</a></li>
|
||||
<li><a href="#amr-0-4-0">0.4.0</a></li>
|
||||
<li><a href="#amr-0-3-0">0.3.0</a></li>
|
||||
<li><a href="#amr-0-2-0">0.2.0</a></li>
|
||||
<li><a href="#amr-0-1-1">0.1.1</a></li>
|
||||
<li><a href="#amr-0-1-0">0.1.0</a></li>
|
||||
</ul>
|
||||
</div>
|
||||
</div>
|
||||
|
||||
@ -1,4 +1,4 @@
|
||||
pandoc: '2.6'
|
||||
pandoc: 2.3.1
|
||||
pkgdown: 1.3.0
|
||||
pkgdown_sha: ~
|
||||
articles:
|
||||
@ -8,7 +8,6 @@ articles:
|
||||
SPSS: SPSS.html
|
||||
WHONET: WHONET.html
|
||||
benchmarks: benchmarks.html
|
||||
freq: freq.html
|
||||
resistance_predict: resistance_predict.html
|
||||
urls:
|
||||
reference: https://msberends.gitlab.io/AMR/reference
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9007</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -1,337 +0,0 @@
|
||||
<!-- Generated by pkgdown: do not edit by hand -->
|
||||
<!DOCTYPE html>
|
||||
<html lang="en">
|
||||
<head>
|
||||
<meta charset="utf-8">
|
||||
<meta http-equiv="X-UA-Compatible" content="IE=edge">
|
||||
<meta name="viewport" content="width=device-width, initial-scale=1.0">
|
||||
|
||||
<title>ITIS: Integrated Taxonomic Information System — ITIS • AMR (for R)</title>
|
||||
|
||||
<!-- favicons -->
|
||||
<link rel="icon" type="image/png" sizes="16x16" href="../favicon-16x16.png">
|
||||
<link rel="icon" type="image/png" sizes="32x32" href="../favicon-32x32.png">
|
||||
<link rel="apple-touch-icon" type="image/png" sizes="180x180" href="../apple-touch-icon.png" />
|
||||
<link rel="apple-touch-icon" type="image/png" sizes="120x120" href="../apple-touch-icon-120x120.png" />
|
||||
<link rel="apple-touch-icon" type="image/png" sizes="76x76" href="../apple-touch-icon-76x76.png" />
|
||||
<link rel="apple-touch-icon" type="image/png" sizes="60x60" href="../apple-touch-icon-60x60.png" />
|
||||
<!-- jquery -->
|
||||
<script src="https://cdnjs.cloudflare.com/ajax/libs/jquery/3.3.1/jquery.min.js" integrity="sha256-FgpCb/KJQlLNfOu91ta32o/NMZxltwRo8QtmkMRdAu8=" crossorigin="anonymous"></script>
|
||||
<!-- Bootstrap -->
|
||||
<link href="https://cdnjs.cloudflare.com/ajax/libs/bootswatch/3.3.7/flatly/bootstrap.min.css" rel="stylesheet" crossorigin="anonymous" />
|
||||
|
||||
<script src="https://cdnjs.cloudflare.com/ajax/libs/twitter-bootstrap/3.3.7/js/bootstrap.min.js" integrity="sha256-U5ZEeKfGNOja007MMD3YBI0A3OSZOQbeG6z2f2Y0hu8=" crossorigin="anonymous"></script>
|
||||
|
||||
<!-- Font Awesome icons -->
|
||||
<link rel="stylesheet" href="https://cdnjs.cloudflare.com/ajax/libs/font-awesome/4.7.0/css/font-awesome.min.css" integrity="sha256-eZrrJcwDc/3uDhsdt61sL2oOBY362qM3lon1gyExkL0=" crossorigin="anonymous" />
|
||||
|
||||
<!-- clipboard.js -->
|
||||
<script src="https://cdnjs.cloudflare.com/ajax/libs/clipboard.js/2.0.4/clipboard.min.js" integrity="sha256-FiZwavyI2V6+EXO1U+xzLG3IKldpiTFf3153ea9zikQ=" crossorigin="anonymous"></script>
|
||||
|
||||
<!-- sticky kit -->
|
||||
<script src="https://cdnjs.cloudflare.com/ajax/libs/sticky-kit/1.1.3/sticky-kit.min.js" integrity="sha256-c4Rlo1ZozqTPE2RLuvbusY3+SU1pQaJC0TjuhygMipw=" crossorigin="anonymous"></script>
|
||||
|
||||
<!-- pkgdown -->
|
||||
<link href="../pkgdown.css" rel="stylesheet">
|
||||
<script src="../pkgdown.js"></script>
|
||||
|
||||
|
||||
<!-- docsearch -->
|
||||
<script src="../docsearch.js"></script>
|
||||
<link rel="stylesheet" href="https://cdnjs.cloudflare.com/ajax/libs/docsearch.js/2.6.1/docsearch.min.css" integrity="sha256-QOSRU/ra9ActyXkIBbiIB144aDBdtvXBcNc3OTNuX/Q=" crossorigin="anonymous" />
|
||||
<link href="../docsearch.css" rel="stylesheet">
|
||||
<script src="https://cdnjs.cloudflare.com/ajax/libs/mark.js/8.11.1/jquery.mark.min.js" integrity="sha256-4HLtjeVgH0eIB3aZ9mLYF6E8oU5chNdjU6p6rrXpl9U=" crossorigin="anonymous"></script>
|
||||
|
||||
|
||||
<link href="../extra.css" rel="stylesheet">
|
||||
<script src="../extra.js"></script>
|
||||
<meta property="og:title" content="ITIS: Integrated Taxonomic Information System — ITIS" />
|
||||
|
||||
<meta property="og:description" content="All taxonomic names of all microorganisms are included in this package, using the authoritative Integrated Taxonomic Information System (ITIS)." />
|
||||
|
||||
<meta property="og:image" content="https://msberends.gitlab.io/AMR/logo.png" />
|
||||
<meta name="twitter:card" content="summary" />
|
||||
|
||||
|
||||
|
||||
<!-- mathjax -->
|
||||
<script src="https://cdnjs.cloudflare.com/ajax/libs/mathjax/2.7.5/MathJax.js" integrity="sha256-nvJJv9wWKEm88qvoQl9ekL2J+k/RWIsaSScxxlsrv8k=" crossorigin="anonymous"></script>
|
||||
<script src="https://cdnjs.cloudflare.com/ajax/libs/mathjax/2.7.5/config/TeX-AMS-MML_HTMLorMML.js" integrity="sha256-84DKXVJXs0/F8OTMzX4UR909+jtl4G7SPypPavF+GfA=" crossorigin="anonymous"></script>
|
||||
|
||||
<!--[if lt IE 9]>
|
||||
<script src="https://oss.maxcdn.com/html5shiv/3.7.3/html5shiv.min.js"></script>
|
||||
<script src="https://oss.maxcdn.com/respond/1.4.2/respond.min.js"></script>
|
||||
<![endif]-->
|
||||
|
||||
|
||||
</head>
|
||||
|
||||
<body>
|
||||
<div class="container template-reference-topic">
|
||||
<header>
|
||||
<div class="navbar navbar-default navbar-fixed-top" role="navigation">
|
||||
<div class="container">
|
||||
<div class="navbar-header">
|
||||
<button type="button" class="navbar-toggle collapsed" data-toggle="collapse" data-target="#navbar" aria-expanded="false">
|
||||
<span class="sr-only">Toggle navigation</span>
|
||||
<span class="icon-bar"></span>
|
||||
<span class="icon-bar"></span>
|
||||
<span class="icon-bar"></span>
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Released version">0.5.0.9018</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
<div id="navbar" class="navbar-collapse collapse">
|
||||
<ul class="nav navbar-nav">
|
||||
<li>
|
||||
<a href="../index.html">
|
||||
<span class="fa fa-home"></span>
|
||||
|
||||
Home
|
||||
</a>
|
||||
</li>
|
||||
<li class="dropdown">
|
||||
<a href="#" class="dropdown-toggle" data-toggle="dropdown" role="button" aria-expanded="false">
|
||||
<span class="fa fa-question-circle"></span>
|
||||
|
||||
How to
|
||||
|
||||
<span class="caret"></span>
|
||||
</a>
|
||||
<ul class="dropdown-menu" role="menu">
|
||||
<li>
|
||||
<a href="../articles/AMR.html">
|
||||
<span class="fa fa-directions"></span>
|
||||
|
||||
Conduct AMR analysis
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/resistance_predict.html">
|
||||
<span class="fa fa-dice"></span>
|
||||
|
||||
Predict antimicrobial resistance
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/WHONET.html">
|
||||
<span class="fa fa-globe-americas"></span>
|
||||
|
||||
Work with WHONET data
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/SPSS.html">
|
||||
<span class="fa fa-file-upload"></span>
|
||||
|
||||
Import data from SPSS/SAS/Stata
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/EUCAST.html">
|
||||
<span class="fa fa-exchange-alt"></span>
|
||||
|
||||
Apply EUCAST rules
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../reference/mo_property.html">
|
||||
<span class="fa fa-bug"></span>
|
||||
|
||||
Get properties of a microorganism
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../reference/atc_property.html">
|
||||
<span class="fa fa-capsules"></span>
|
||||
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/G_test.html">
|
||||
<span class="fa fa-clipboard-check"></span>
|
||||
|
||||
Use the G-test
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
Other: benchmarks
|
||||
</a>
|
||||
</li>
|
||||
</ul>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../reference/">
|
||||
<span class="fa fa-book-open"></span>
|
||||
|
||||
Manual
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../authors.html">
|
||||
<span class="fa fa-users"></span>
|
||||
|
||||
Authors
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../news/">
|
||||
<span class="far fa far fa-newspaper"></span>
|
||||
|
||||
Changelog
|
||||
</a>
|
||||
</li>
|
||||
</ul>
|
||||
|
||||
<ul class="nav navbar-nav navbar-right">
|
||||
<li>
|
||||
<a href="https://gitlab.com/msberends/AMR">
|
||||
<span class="fab fa fab fa-gitlab"></span>
|
||||
|
||||
Source Code
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../LICENSE-text.html">
|
||||
<span class="fa fa-book"></span>
|
||||
|
||||
Licence
|
||||
</a>
|
||||
</li>
|
||||
</ul>
|
||||
|
||||
<form class="navbar-form navbar-right" role="search">
|
||||
<div class="form-group">
|
||||
<input type="search" class="form-control" name="search-input" id="search-input" placeholder="Search..." aria-label="Search for..." autocomplete="off">
|
||||
</div>
|
||||
</form>
|
||||
|
||||
</div><!--/.nav-collapse -->
|
||||
</div><!--/.container -->
|
||||
</div><!--/.navbar -->
|
||||
|
||||
|
||||
</header>
|
||||
|
||||
<div class="row">
|
||||
<div class="col-md-9 contents">
|
||||
<div class="page-header">
|
||||
<h1>ITIS: Integrated Taxonomic Information System</h1>
|
||||
|
||||
<div class="hidden name"><code>ITIS.Rd</code></div>
|
||||
</div>
|
||||
|
||||
<div class="ref-description">
|
||||
|
||||
<p>All taxonomic names of all microorganisms are included in this package, using the authoritative Integrated Taxonomic Information System (ITIS).</p>
|
||||
|
||||
</div>
|
||||
|
||||
|
||||
<h2 class="hasAnchor" id="itis"><a class="anchor" href="#itis"></a>ITIS</h2>
|
||||
|
||||
|
||||
<p><img src='figures/logo_itis.jpg' height=60px style=margin-bottom:5px /> <br />
|
||||
This package contains the <strong>complete microbial taxonomic data</strong> (with all nine taxonomic ranks - from kingdom to subspecies) from the publicly available Integrated Taxonomic Information System (ITIS, <a href='https://www.itis.gov'>https://www.itis.gov</a>).</p>
|
||||
<p>All ~20,000 (sub)species from <strong>the taxonomic kingdoms Bacteria, Fungi and Protozoa are included in this package</strong>, as well as all their ~2,500 previously accepted names known to ITIS. Furthermore, the responsible authors and year of publication are available. This allows users to use authoritative taxonomic information for their data analysis on any microorganism, not only human pathogens. It also helps to quickly determine the Gram stain of bacteria, since ITIS honours the taxonomic branching order of bacterial phyla according to Cavalier-Smith (2002), which defines that all bacteria are classified into either subkingdom Negibacteria or subkingdom Posibacteria.</p>
|
||||
<p>ITIS is a partnership of U.S., Canadian, and Mexican agencies and taxonomic specialists [3].</p>
|
||||
|
||||
<h2 class="hasAnchor" id="read-more-on-our-website-"><a class="anchor" href="#read-more-on-our-website-"></a>Read more on our website!</h2>
|
||||
|
||||
|
||||
<p><img src='figures/logo.png' height=40px style=margin-bottom:5px /> <br />
|
||||
On our website <a href='https://msberends.gitlab.io/AMR'>https://msberends.gitlab.io/AMR</a> you can find <a href='https://msberends.gitlab.io/AMR/articles/AMR.html'>a comprehensive tutorial</a> about how to conduct AMR analysis, the <a href='https://msberends.gitlab.io/AMR/reference'>complete documentation of all functions</a> (which reads a lot easier than here in R) and <a href='https://msberends.gitlab.io/AMR/articles/WHONET.html'>an example analysis using WHONET data</a>.</p>
|
||||
|
||||
|
||||
<h2 class="hasAnchor" id="examples"><a class="anchor" href="#examples"></a>Examples</h2>
|
||||
<pre class="examples"><span class='co'># NOT RUN {</span>
|
||||
<span class='co'># Get a note when a species was renamed</span>
|
||||
<span class='fu'><a href='mo_property.html'>mo_shortname</a></span>(<span class='st'>"Chlamydia psittaci"</span>)
|
||||
<span class='co'># Note: 'Chlamydia psittaci' (Page, 1968) was renamed</span>
|
||||
<span class='co'># 'Chlamydophila psittaci' (Everett et al., 1999)</span>
|
||||
<span class='co'># [1] "C. psittaci"</span>
|
||||
|
||||
<span class='co'># Get any property from the entire taxonomic tree for all included species</span>
|
||||
<span class='fu'><a href='mo_property.html'>mo_class</a></span>(<span class='st'>"E. coli"</span>)
|
||||
<span class='co'># [1] "Gammaproteobacteria"</span>
|
||||
|
||||
<span class='fu'><a href='mo_property.html'>mo_family</a></span>(<span class='st'>"E. coli"</span>)
|
||||
<span class='co'># [1] "Enterobacteriaceae"</span>
|
||||
|
||||
<span class='fu'><a href='mo_property.html'>mo_subkingdom</a></span>(<span class='st'>"E. coli"</span>)
|
||||
<span class='co'># [1] "Negibacteria"</span>
|
||||
|
||||
<span class='fu'><a href='mo_property.html'>mo_gramstain</a></span>(<span class='st'>"E. coli"</span>) <span class='co'># based on subkingdom</span>
|
||||
<span class='co'># [1] "Gram negative"</span>
|
||||
|
||||
<span class='fu'><a href='mo_property.html'>mo_ref</a></span>(<span class='st'>"E. coli"</span>)
|
||||
<span class='co'># [1] "Castellani and Chalmers, 1919"</span>
|
||||
|
||||
<span class='co'># Do not get mistaken - the package only includes microorganisms</span>
|
||||
<span class='fu'><a href='mo_property.html'>mo_phylum</a></span>(<span class='st'>"C. elegans"</span>)
|
||||
<span class='co'># [1] "Cyanobacteria" # Bacteria?!</span>
|
||||
<span class='fu'><a href='mo_property.html'>mo_fullname</a></span>(<span class='st'>"C. elegans"</span>)
|
||||
<span class='co'># [1] "Chroococcus limneticus elegans" # Because a microorganism was found</span>
|
||||
<span class='co'># }</span></pre>
|
||||
</div>
|
||||
<div class="col-md-3 hidden-xs hidden-sm" id="sidebar">
|
||||
<h2>Contents</h2>
|
||||
<ul class="nav nav-pills nav-stacked">
|
||||
|
||||
<li><a href="#itis">ITIS</a></li>
|
||||
|
||||
<li><a href="#read-more-on-our-website-">Read more on our website!</a></li>
|
||||
|
||||
<li><a href="#examples">Examples</a></li>
|
||||
</ul>
|
||||
|
||||
</div>
|
||||
</div>
|
||||
|
||||
<footer>
|
||||
<div class="copyright">
|
||||
<p>Developed by <a href='https://www.rug.nl/staff/m.s.berends/'>Matthijs S. Berends</a>, <a href='https://www.rug.nl/staff/c.f.luz/'>Christian F. Luz</a>, <a href='https://www.rug.nl/staff/c.glasner/'>Corinna Glasner</a>, <a href='https://www.rug.nl/staff/a.w.friedrich/'>Alex W. Friedrich</a>, <a href='https://www.rug.nl/staff/b.sinha/'>Bhanu N. M. Sinha</a>.</p>
|
||||
</div>
|
||||
|
||||
<div class="pkgdown">
|
||||
<p>Site built with <a href="https://pkgdown.r-lib.org/">pkgdown</a> 1.3.0.</p>
|
||||
</div>
|
||||
</footer>
|
||||
</div>
|
||||
|
||||
|
||||
<script src="https://cdnjs.cloudflare.com/ajax/libs/docsearch.js/2.6.1/docsearch.min.js" integrity="sha256-GKvGqXDznoRYHCwKXGnuchvKSwmx9SRMrZOTh2g4Sb0=" crossorigin="anonymous"></script>
|
||||
<script>
|
||||
docsearch({
|
||||
|
||||
|
||||
apiKey: 'f737050abfd4d726c63938e18f8c496e',
|
||||
indexName: 'amr',
|
||||
inputSelector: 'input#search-input.form-control',
|
||||
transformData: function(hits) {
|
||||
return hits.map(function (hit) {
|
||||
hit.url = updateHitURL(hit);
|
||||
return hit;
|
||||
});
|
||||
}
|
||||
});
|
||||
</script>
|
||||
|
||||
|
||||
</body>
|
||||
</html>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9026</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -246,7 +239,7 @@
|
||||
|
||||
|
||||
<p><img src='figures/logo_who.png' height=60px style=margin-bottom:5px /> <br />
|
||||
This package contains <strong>all ~450 antimicrobial drugs</strong> and their Anatomical Therapeutic Chemical (ATC) codes, ATC groups and Defined Daily Dose (DDD) from the World Health Organization Collaborating Centre for Drug Statistics Methodology (WHOCC, <a href='https://www.whocc.no'>https://www.whocc.no</a>) and the Pharmaceuticals Community Register of the European Commission (<a href='http://ec.europa.eu/health/documents/community-register/html/atc.htm'>http://ec.europa.eu/health/documents/community-register/html/atc.htm</a>).</p>
|
||||
This package contains <strong>all ~450 antimicrobial drugs</strong> and their Anatomical Therapeutic Chemical (ATC) codes, ATC groups and Defined Daily Dose (DDD) from the World Health Organization Collaborating Centre for Drug Statistics Methodology (WHOCC, <a href='https://www.whocc.no'>https://www.whocc.no</a>) and the Pharmaceuticals Community Register of the European Commission (<a href='http://ec.europa.eu/health/documents/community-register/html/atc.htm'>http://ec.europa.eu/health/documents/community-register/html/atc.htm</a>). <strong>NOTE: The WHOCC copyright does not allow use for commercial purposes, unlike any other info from this package. See <a href='https://www.whocc.no/copyright_disclaimer/'>https://www.whocc.no/copyright_disclaimer/</a>.</strong></p>
|
||||
<p>These have become the gold standard for international drug utilisation monitoring and research.</p>
|
||||
<p>The WHOCC is located in Oslo at the Norwegian Institute of Public Health and funded by the Norwegian government. The European Commission is the executive of the European Union and promotes its general interest.</p>
|
||||
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9007</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9026</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -276,7 +269,7 @@
|
||||
|
||||
|
||||
<p><img src='figures/logo_who.png' height=60px style=margin-bottom:5px /> <br />
|
||||
This package contains <strong>all ~450 antimicrobial drugs</strong> and their Anatomical Therapeutic Chemical (ATC) codes, ATC groups and Defined Daily Dose (DDD) from the World Health Organization Collaborating Centre for Drug Statistics Methodology (WHOCC, <a href='https://www.whocc.no'>https://www.whocc.no</a>) and the Pharmaceuticals Community Register of the European Commission (<a href='http://ec.europa.eu/health/documents/community-register/html/atc.htm'>http://ec.europa.eu/health/documents/community-register/html/atc.htm</a>).</p>
|
||||
This package contains <strong>all ~450 antimicrobial drugs</strong> and their Anatomical Therapeutic Chemical (ATC) codes, ATC groups and Defined Daily Dose (DDD) from the World Health Organization Collaborating Centre for Drug Statistics Methodology (WHOCC, <a href='https://www.whocc.no'>https://www.whocc.no</a>) and the Pharmaceuticals Community Register of the European Commission (<a href='http://ec.europa.eu/health/documents/community-register/html/atc.htm'>http://ec.europa.eu/health/documents/community-register/html/atc.htm</a>). <strong>NOTE: The WHOCC copyright does not allow use for commercial purposes, unlike any other info from this package. See <a href='https://www.whocc.no/copyright_disclaimer/'>https://www.whocc.no/copyright_disclaimer/</a>.</strong></p>
|
||||
<p>These have become the gold standard for international drug utilisation monitoring and research.</p>
|
||||
<p>The WHOCC is located in Oslo at the Norwegian Institute of Public Health and funded by the Norwegian government. The European Commission is the executive of the European Union and promotes its general interest.</p>
|
||||
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9026</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -274,7 +267,7 @@
|
||||
|
||||
|
||||
<p><img src='figures/logo_who.png' height=60px style=margin-bottom:5px /> <br />
|
||||
This package contains <strong>all ~450 antimicrobial drugs</strong> and their Anatomical Therapeutic Chemical (ATC) codes, ATC groups and Defined Daily Dose (DDD) from the World Health Organization Collaborating Centre for Drug Statistics Methodology (WHOCC, <a href='https://www.whocc.no'>https://www.whocc.no</a>) and the Pharmaceuticals Community Register of the European Commission (<a href='http://ec.europa.eu/health/documents/community-register/html/atc.htm'>http://ec.europa.eu/health/documents/community-register/html/atc.htm</a>).</p>
|
||||
This package contains <strong>all ~450 antimicrobial drugs</strong> and their Anatomical Therapeutic Chemical (ATC) codes, ATC groups and Defined Daily Dose (DDD) from the World Health Organization Collaborating Centre for Drug Statistics Methodology (WHOCC, <a href='https://www.whocc.no'>https://www.whocc.no</a>) and the Pharmaceuticals Community Register of the European Commission (<a href='http://ec.europa.eu/health/documents/community-register/html/atc.htm'>http://ec.europa.eu/health/documents/community-register/html/atc.htm</a>). <strong>NOTE: The WHOCC copyright does not allow use for commercial purposes, unlike any other info from this package. See <a href='https://www.whocc.no/copyright_disclaimer/'>https://www.whocc.no/copyright_disclaimer/</a>.</strong></p>
|
||||
<p>These have become the gold standard for international drug utilisation monitoring and research.</p>
|
||||
<p>The WHOCC is located in Oslo at the Norwegian Institute of Public Health and funded by the Norwegian government. The European Commission is the executive of the European Union and promotes its general interest.</p>
|
||||
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9026</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -296,7 +289,9 @@
|
||||
|
||||
<span class='fu'><a href='https://www.rdocumentation.org/packages/graphics/topics/plot'>plot</a></span>(<span class='no'>mic_data</span>)
|
||||
<span class='fu'><a href='https://www.rdocumentation.org/packages/graphics/topics/barplot'>barplot</a></span>(<span class='no'>mic_data</span>)
|
||||
<span class='fu'><a href='freq.html'>freq</a></span>(<span class='no'>mic_data</span>)
|
||||
|
||||
<span class='fu'><a href='https://www.rdocumentation.org/packages/base/topics/library'>library</a></span>(<span class='no'>clean</span>)
|
||||
<span class='fu'><a href='https://www.rdocumentation.org/packages/clean/topics/freq'>freq</a></span>(<span class='no'>mic_data</span>)
|
||||
<span class='co'># }</span></pre>
|
||||
</div>
|
||||
<div class="col-md-3 hidden-xs hidden-sm" id="sidebar">
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9007</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9007</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9026</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -343,7 +336,9 @@
|
||||
|
||||
<span class='fu'><a href='https://www.rdocumentation.org/packages/graphics/topics/plot'>plot</a></span>(<span class='no'>rsi_data</span>) <span class='co'># for percentages</span>
|
||||
<span class='fu'><a href='https://www.rdocumentation.org/packages/graphics/topics/barplot'>barplot</a></span>(<span class='no'>rsi_data</span>) <span class='co'># for frequencies</span>
|
||||
<span class='fu'><a href='freq.html'>freq</a></span>(<span class='no'>rsi_data</span>) <span class='co'># frequency table with informative header</span>
|
||||
|
||||
<span class='fu'><a href='https://www.rdocumentation.org/packages/base/topics/library'>library</a></span>(<span class='no'>clean</span>)
|
||||
<span class='fu'><a href='https://www.rdocumentation.org/packages/clean/topics/freq'>freq</a></span>(<span class='no'>rsi_data</span>) <span class='co'># frequency table with informative header</span>
|
||||
|
||||
<span class='co'># using dplyr's mutate</span>
|
||||
<span class='fu'><a href='https://www.rdocumentation.org/packages/base/topics/library'>library</a></span>(<span class='no'>dplyr</span>)
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9026</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -271,8 +264,9 @@ This package contains the complete taxonomic tree of almost all microorganisms (
|
||||
<h2 class="hasAnchor" id="examples"><a class="anchor" href="#examples"></a>Examples</h2>
|
||||
<pre class="examples"><span class='co'># NOT RUN {</span>
|
||||
<span class='fu'><a href='https://www.rdocumentation.org/packages/base/topics/library'>library</a></span>(<span class='no'>dplyr</span>)
|
||||
<span class='no'>microorganisms</span> <span class='kw'>%>%</span> <span class='fu'><a href='freq.html'>freq</a></span>(<span class='no'>kingdom</span>)
|
||||
<span class='no'>microorganisms</span> <span class='kw'>%>%</span> <span class='fu'><a href='https://dplyr.tidyverse.org/reference/group_by.html'>group_by</a></span>(<span class='no'>kingdom</span>) <span class='kw'>%>%</span> <span class='fu'><a href='freq.html'>freq</a></span>(<span class='no'>phylum</span>, <span class='kw'>nmax</span> <span class='kw'>=</span> <span class='kw'>NULL</span>)
|
||||
<span class='fu'><a href='https://www.rdocumentation.org/packages/base/topics/library'>library</a></span>(<span class='no'>clean</span>)
|
||||
<span class='no'>microorganisms</span> <span class='kw'>%>%</span> <span class='fu'><a href='https://www.rdocumentation.org/packages/clean/topics/freq'>freq</a></span>(<span class='no'>kingdom</span>)
|
||||
<span class='no'>microorganisms</span> <span class='kw'>%>%</span> <span class='fu'><a href='https://dplyr.tidyverse.org/reference/group_by.html'>group_by</a></span>(<span class='no'>kingdom</span>) <span class='kw'>%>%</span> <span class='fu'><a href='https://www.rdocumentation.org/packages/clean/topics/freq'>freq</a></span>(<span class='no'>phylum</span>, <span class='kw'>nmax</span> <span class='kw'>=</span> <span class='kw'>NULL</span>)
|
||||
<span class='co'># }</span></pre>
|
||||
</div>
|
||||
<div class="col-md-3 hidden-xs hidden-sm" id="sidebar">
|
||||
|
||||
@ -81,7 +81,7 @@ count_R and count_IR can be used to count resistant isolates, count_S and count_
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9007</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -159,13 +159,6 @@ count_R and count_IR can be used to count resistant isolates, count_S and count_
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9026</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -265,7 +258,7 @@
|
||||
</tr>
|
||||
<tr>
|
||||
<th>verbose</th>
|
||||
<td><p>a logical to indicate whether extensive info should be returned as a <code>data.frame</code> with info about which rows and columns are effected. It runs all EUCAST rules, but will not be applied to an output - only an informative <code>data.frame</code> with changes will be returned as output.</p></td>
|
||||
<td><p>a logical to turn Verbose mode on and off (default is off). In Verbose mode, the function does not apply rules to the data, but instead returns a <code>data.frame</code> with extensive info about which rows and columns would be effected and in which way.</p></td>
|
||||
</tr>
|
||||
<tr>
|
||||
<th>...</th>
|
||||
@ -381,8 +374,6 @@
|
||||
|
||||
<h2 class="hasAnchor" id="examples"><a class="anchor" href="#examples"></a>Examples</h2>
|
||||
<pre class="examples"><span class='co'># NOT RUN {</span>
|
||||
<span class='no'>a</span> <span class='kw'><-</span> <span class='fu'>eucast_rules</span>(<span class='no'>septic_patients</span>)
|
||||
|
||||
<span class='no'>a</span> <span class='kw'><-</span> <span class='fu'><a href='https://www.rdocumentation.org/packages/base/topics/data.frame'>data.frame</a></span>(<span class='kw'>mo</span> <span class='kw'>=</span> <span class='fu'><a href='https://www.rdocumentation.org/packages/base/topics/c'>c</a></span>(<span class='st'>"Staphylococcus aureus"</span>,
|
||||
<span class='st'>"Enterococcus faecalis"</span>,
|
||||
<span class='st'>"Escherichia coli"</span>,
|
||||
@ -418,7 +409,7 @@
|
||||
<span class='co'># 5 Pseudomonas aeruginosa R R - - R R R</span>
|
||||
|
||||
|
||||
<span class='co'># do not apply EUCAST rules, but rather get a a data.frame</span>
|
||||
<span class='co'># do not apply EUCAST rules, but rather get a data.frame</span>
|
||||
<span class='co'># with 18 rows, containing all details about the transformations:</span>
|
||||
<span class='no'>c</span> <span class='kw'><-</span> <span class='fu'>eucast_rules</span>(<span class='no'>a</span>, <span class='kw'>verbose</span> <span class='kw'>=</span> <span class='fl'>TRUE</span>)
|
||||
<span class='co'># }</span></pre>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -81,7 +81,7 @@ top_freq can be used to get the top/bottom n items of a frequency table, with co
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9007</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9009</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9007</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -457,7 +450,7 @@
|
||||
<span class='co'># create new bacterial ID's, with all CoNS under the same group (Becker et al.)</span>
|
||||
<span class='fu'><a href='https://dplyr.tidyverse.org/reference/mutate.html'>mutate</a></span>(<span class='kw'>mo</span> <span class='kw'>=</span> <span class='fu'><a href='as.mo.html'>as.mo</a></span>(<span class='no'>mo</span>, <span class='kw'>Becker</span> <span class='kw'>=</span> <span class='fl'>TRUE</span>)) <span class='kw'>%>%</span>
|
||||
<span class='co'># filter on top three bacterial ID's</span>
|
||||
<span class='fu'><a href='https://dplyr.tidyverse.org/reference/filter.html'>filter</a></span>(<span class='no'>mo</span> <span class='kw'>%in%</span> <span class='fu'><a href='freq.html'>top_freq</a></span>(<span class='fu'><a href='freq.html'>freq</a></span>(<span class='no'>.</span>$<span class='no'>mo</span>), <span class='fl'>3</span>)) <span class='kw'>%>%</span>
|
||||
<span class='fu'><a href='https://dplyr.tidyverse.org/reference/filter.html'>filter</a></span>(<span class='no'>mo</span> <span class='kw'>%in%</span> <span class='fu'>top_freq</span>(<span class='fu'>freq</span>(<span class='no'>.</span>$<span class='no'>mo</span>), <span class='fl'>3</span>)) <span class='kw'>%>%</span>
|
||||
<span class='co'># filter on first isolates</span>
|
||||
<span class='fu'><a href='first_isolate.html'>filter_first_isolate</a></span>() <span class='kw'>%>%</span>
|
||||
<span class='co'># get short MO names (like "E. coli")</span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -78,7 +78,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9008</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9026</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -156,13 +156,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -411,7 +404,7 @@
|
||||
<tr>
|
||||
<th colspan="2">
|
||||
<h2 id="section-analysing-your-data" class="hasAnchor"><a href="#section-analysing-your-data" class="anchor"></a>Analysing your data</h2>
|
||||
<p class="section-desc"><p>Functions for conducting AMR analysis, like counting isolates, calculating resistance or susceptibility, creating frequency tables or make plots.</p></p>
|
||||
<p class="section-desc"><p>Functions for conducting AMR analysis, like counting isolates, calculating resistance or susceptibility, or make plots.</p></p>
|
||||
</th>
|
||||
</tr>
|
||||
<tr>
|
||||
@ -440,12 +433,6 @@
|
||||
<td><p>Filter isolates on result in antibiotic class</p></td>
|
||||
</tr><tr>
|
||||
|
||||
<td>
|
||||
<p><code><a href="freq.html">freq()</a></code> <code><a href="freq.html">frequency_tbl()</a></code> <code><a href="freq.html">top_freq()</a></code> <code><a href="freq.html">header()</a></code> <code><a href="freq.html">print(<i><freq></i>)</a></code> </p>
|
||||
</td>
|
||||
<td><p>Frequency table</p></td>
|
||||
</tr><tr>
|
||||
|
||||
<td>
|
||||
<p><code><a href="g.test.html">g.test()</a></code> </p>
|
||||
</td>
|
||||
|
||||
@ -1,337 +0,0 @@
|
||||
<!-- Generated by pkgdown: do not edit by hand -->
|
||||
<!DOCTYPE html>
|
||||
<html lang="en">
|
||||
<head>
|
||||
<meta charset="utf-8">
|
||||
<meta http-equiv="X-UA-Compatible" content="IE=edge">
|
||||
<meta name="viewport" content="width=device-width, initial-scale=1.0">
|
||||
|
||||
<title>ITIS: Integrated Taxonomic Information System — ITIS • AMR (for R)</title>
|
||||
|
||||
<!-- favicons -->
|
||||
<link rel="icon" type="image/png" sizes="16x16" href="../favicon-16x16.png">
|
||||
<link rel="icon" type="image/png" sizes="32x32" href="../favicon-32x32.png">
|
||||
<link rel="apple-touch-icon" type="image/png" sizes="180x180" href="../apple-touch-icon.png" />
|
||||
<link rel="apple-touch-icon" type="image/png" sizes="120x120" href="../apple-touch-icon-120x120.png" />
|
||||
<link rel="apple-touch-icon" type="image/png" sizes="76x76" href="../apple-touch-icon-76x76.png" />
|
||||
<link rel="apple-touch-icon" type="image/png" sizes="60x60" href="../apple-touch-icon-60x60.png" />
|
||||
<!-- jquery -->
|
||||
<script src="https://cdnjs.cloudflare.com/ajax/libs/jquery/3.3.1/jquery.min.js" integrity="sha256-FgpCb/KJQlLNfOu91ta32o/NMZxltwRo8QtmkMRdAu8=" crossorigin="anonymous"></script>
|
||||
<!-- Bootstrap -->
|
||||
<link href="https://cdnjs.cloudflare.com/ajax/libs/bootswatch/3.3.7/flatly/bootstrap.min.css" rel="stylesheet" crossorigin="anonymous" />
|
||||
|
||||
<script src="https://cdnjs.cloudflare.com/ajax/libs/twitter-bootstrap/3.3.7/js/bootstrap.min.js" integrity="sha256-U5ZEeKfGNOja007MMD3YBI0A3OSZOQbeG6z2f2Y0hu8=" crossorigin="anonymous"></script>
|
||||
|
||||
<!-- Font Awesome icons -->
|
||||
<link rel="stylesheet" href="https://cdnjs.cloudflare.com/ajax/libs/font-awesome/4.7.0/css/font-awesome.min.css" integrity="sha256-eZrrJcwDc/3uDhsdt61sL2oOBY362qM3lon1gyExkL0=" crossorigin="anonymous" />
|
||||
|
||||
<!-- clipboard.js -->
|
||||
<script src="https://cdnjs.cloudflare.com/ajax/libs/clipboard.js/2.0.4/clipboard.min.js" integrity="sha256-FiZwavyI2V6+EXO1U+xzLG3IKldpiTFf3153ea9zikQ=" crossorigin="anonymous"></script>
|
||||
|
||||
<!-- sticky kit -->
|
||||
<script src="https://cdnjs.cloudflare.com/ajax/libs/sticky-kit/1.1.3/sticky-kit.min.js" integrity="sha256-c4Rlo1ZozqTPE2RLuvbusY3+SU1pQaJC0TjuhygMipw=" crossorigin="anonymous"></script>
|
||||
|
||||
<!-- pkgdown -->
|
||||
<link href="../pkgdown.css" rel="stylesheet">
|
||||
<script src="../pkgdown.js"></script>
|
||||
|
||||
|
||||
<!-- docsearch -->
|
||||
<script src="../docsearch.js"></script>
|
||||
<link rel="stylesheet" href="https://cdnjs.cloudflare.com/ajax/libs/docsearch.js/2.6.1/docsearch.min.css" integrity="sha256-QOSRU/ra9ActyXkIBbiIB144aDBdtvXBcNc3OTNuX/Q=" crossorigin="anonymous" />
|
||||
<link href="../docsearch.css" rel="stylesheet">
|
||||
<script src="https://cdnjs.cloudflare.com/ajax/libs/mark.js/8.11.1/jquery.mark.min.js" integrity="sha256-4HLtjeVgH0eIB3aZ9mLYF6E8oU5chNdjU6p6rrXpl9U=" crossorigin="anonymous"></script>
|
||||
|
||||
|
||||
<link href="../extra.css" rel="stylesheet">
|
||||
<script src="../extra.js"></script>
|
||||
<meta property="og:title" content="ITIS: Integrated Taxonomic Information System — ITIS" />
|
||||
|
||||
<meta property="og:description" content="All taxonomic names of all microorganisms are included in this package, using the authoritative Integrated Taxonomic Information System (ITIS)." />
|
||||
|
||||
<meta property="og:image" content="https://msberends.gitlab.io/AMR/logo.png" />
|
||||
<meta name="twitter:card" content="summary" />
|
||||
|
||||
|
||||
|
||||
<!-- mathjax -->
|
||||
<script src="https://cdnjs.cloudflare.com/ajax/libs/mathjax/2.7.5/MathJax.js" integrity="sha256-nvJJv9wWKEm88qvoQl9ekL2J+k/RWIsaSScxxlsrv8k=" crossorigin="anonymous"></script>
|
||||
<script src="https://cdnjs.cloudflare.com/ajax/libs/mathjax/2.7.5/config/TeX-AMS-MML_HTMLorMML.js" integrity="sha256-84DKXVJXs0/F8OTMzX4UR909+jtl4G7SPypPavF+GfA=" crossorigin="anonymous"></script>
|
||||
|
||||
<!--[if lt IE 9]>
|
||||
<script src="https://oss.maxcdn.com/html5shiv/3.7.3/html5shiv.min.js"></script>
|
||||
<script src="https://oss.maxcdn.com/respond/1.4.2/respond.min.js"></script>
|
||||
<![endif]-->
|
||||
|
||||
|
||||
</head>
|
||||
|
||||
<body>
|
||||
<div class="container template-reference-topic">
|
||||
<header>
|
||||
<div class="navbar navbar-default navbar-fixed-top" role="navigation">
|
||||
<div class="container">
|
||||
<div class="navbar-header">
|
||||
<button type="button" class="navbar-toggle collapsed" data-toggle="collapse" data-target="#navbar" aria-expanded="false">
|
||||
<span class="sr-only">Toggle navigation</span>
|
||||
<span class="icon-bar"></span>
|
||||
<span class="icon-bar"></span>
|
||||
<span class="icon-bar"></span>
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Released version">0.5.0.9018</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
<div id="navbar" class="navbar-collapse collapse">
|
||||
<ul class="nav navbar-nav">
|
||||
<li>
|
||||
<a href="../index.html">
|
||||
<span class="fa fa-home"></span>
|
||||
|
||||
Home
|
||||
</a>
|
||||
</li>
|
||||
<li class="dropdown">
|
||||
<a href="#" class="dropdown-toggle" data-toggle="dropdown" role="button" aria-expanded="false">
|
||||
<span class="fa fa-question-circle"></span>
|
||||
|
||||
How to
|
||||
|
||||
<span class="caret"></span>
|
||||
</a>
|
||||
<ul class="dropdown-menu" role="menu">
|
||||
<li>
|
||||
<a href="../articles/AMR.html">
|
||||
<span class="fa fa-directions"></span>
|
||||
|
||||
Conduct AMR analysis
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/resistance_predict.html">
|
||||
<span class="fa fa-dice"></span>
|
||||
|
||||
Predict antimicrobial resistance
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/WHONET.html">
|
||||
<span class="fa fa-globe-americas"></span>
|
||||
|
||||
Work with WHONET data
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/SPSS.html">
|
||||
<span class="fa fa-file-upload"></span>
|
||||
|
||||
Import data from SPSS/SAS/Stata
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/EUCAST.html">
|
||||
<span class="fa fa-exchange-alt"></span>
|
||||
|
||||
Apply EUCAST rules
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../reference/mo_property.html">
|
||||
<span class="fa fa-bug"></span>
|
||||
|
||||
Get properties of a microorganism
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../reference/atc_property.html">
|
||||
<span class="fa fa-capsules"></span>
|
||||
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/G_test.html">
|
||||
<span class="fa fa-clipboard-check"></span>
|
||||
|
||||
Use the G-test
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
Other: benchmarks
|
||||
</a>
|
||||
</li>
|
||||
</ul>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../reference/">
|
||||
<span class="fa fa-book-open"></span>
|
||||
|
||||
Manual
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../authors.html">
|
||||
<span class="fa fa-users"></span>
|
||||
|
||||
Authors
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../news/">
|
||||
<span class="far fa far fa-newspaper"></span>
|
||||
|
||||
Changelog
|
||||
</a>
|
||||
</li>
|
||||
</ul>
|
||||
|
||||
<ul class="nav navbar-nav navbar-right">
|
||||
<li>
|
||||
<a href="https://gitlab.com/msberends/AMR">
|
||||
<span class="fab fa fab fa-gitlab"></span>
|
||||
|
||||
Source Code
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../LICENSE-text.html">
|
||||
<span class="fa fa-book"></span>
|
||||
|
||||
Licence
|
||||
</a>
|
||||
</li>
|
||||
</ul>
|
||||
|
||||
<form class="navbar-form navbar-right" role="search">
|
||||
<div class="form-group">
|
||||
<input type="search" class="form-control" name="search-input" id="search-input" placeholder="Search..." aria-label="Search for..." autocomplete="off">
|
||||
</div>
|
||||
</form>
|
||||
|
||||
</div><!--/.nav-collapse -->
|
||||
</div><!--/.container -->
|
||||
</div><!--/.navbar -->
|
||||
|
||||
|
||||
</header>
|
||||
|
||||
<div class="row">
|
||||
<div class="col-md-9 contents">
|
||||
<div class="page-header">
|
||||
<h1>ITIS: Integrated Taxonomic Information System</h1>
|
||||
|
||||
<div class="hidden name"><code>ITIS.Rd</code></div>
|
||||
</div>
|
||||
|
||||
<div class="ref-description">
|
||||
|
||||
<p>All taxonomic names of all microorganisms are included in this package, using the authoritative Integrated Taxonomic Information System (ITIS).</p>
|
||||
|
||||
</div>
|
||||
|
||||
|
||||
<h2 class="hasAnchor" id="itis"><a class="anchor" href="#itis"></a>ITIS</h2>
|
||||
|
||||
|
||||
<p><img src='figures/logo_itis.jpg' height=60px style=margin-bottom:5px /> <br />
|
||||
This package contains the <strong>complete microbial taxonomic data</strong> (with all nine taxonomic ranks - from kingdom to subspecies) from the publicly available Integrated Taxonomic Information System (ITIS, <a href='https://www.itis.gov'>https://www.itis.gov</a>).</p>
|
||||
<p>All ~20,000 (sub)species from <strong>the taxonomic kingdoms Bacteria, Fungi and Protozoa are included in this package</strong>, as well as all their ~2,500 previously accepted names known to ITIS. Furthermore, the responsible authors and year of publication are available. This allows users to use authoritative taxonomic information for their data analysis on any microorganism, not only human pathogens. It also helps to quickly determine the Gram stain of bacteria, since ITIS honours the taxonomic branching order of bacterial phyla according to Cavalier-Smith (2002), which defines that all bacteria are classified into either subkingdom Negibacteria or subkingdom Posibacteria.</p>
|
||||
<p>ITIS is a partnership of U.S., Canadian, and Mexican agencies and taxonomic specialists [3].</p>
|
||||
|
||||
<h2 class="hasAnchor" id="read-more-on-our-website-"><a class="anchor" href="#read-more-on-our-website-"></a>Read more on our website!</h2>
|
||||
|
||||
|
||||
<p><img src='figures/logo.png' height=40px style=margin-bottom:5px /> <br />
|
||||
On our website <a href='https://msberends.gitlab.io/AMR'>https://msberends.gitlab.io/AMR</a> you can find <a href='https://msberends.gitlab.io/AMR/articles/AMR.html'>a comprehensive tutorial</a> about how to conduct AMR analysis, the <a href='https://msberends.gitlab.io/AMR/reference'>complete documentation of all functions</a> (which reads a lot easier than here in R) and <a href='https://msberends.gitlab.io/AMR/articles/WHONET.html'>an example analysis using WHONET data</a>.</p>
|
||||
|
||||
|
||||
<h2 class="hasAnchor" id="examples"><a class="anchor" href="#examples"></a>Examples</h2>
|
||||
<pre class="examples"><span class='co'># NOT RUN {</span>
|
||||
<span class='co'># Get a note when a species was renamed</span>
|
||||
<span class='fu'><a href='mo_property.html'>mo_shortname</a></span>(<span class='st'>"Chlamydia psittaci"</span>)
|
||||
<span class='co'># Note: 'Chlamydia psittaci' (Page, 1968) was renamed</span>
|
||||
<span class='co'># 'Chlamydophila psittaci' (Everett et al., 1999)</span>
|
||||
<span class='co'># [1] "C. psittaci"</span>
|
||||
|
||||
<span class='co'># Get any property from the entire taxonomic tree for all included species</span>
|
||||
<span class='fu'><a href='mo_property.html'>mo_class</a></span>(<span class='st'>"E. coli"</span>)
|
||||
<span class='co'># [1] "Gammaproteobacteria"</span>
|
||||
|
||||
<span class='fu'><a href='mo_property.html'>mo_family</a></span>(<span class='st'>"E. coli"</span>)
|
||||
<span class='co'># [1] "Enterobacteriaceae"</span>
|
||||
|
||||
<span class='fu'><a href='mo_property.html'>mo_subkingdom</a></span>(<span class='st'>"E. coli"</span>)
|
||||
<span class='co'># [1] "Negibacteria"</span>
|
||||
|
||||
<span class='fu'><a href='mo_property.html'>mo_gramstain</a></span>(<span class='st'>"E. coli"</span>) <span class='co'># based on subkingdom</span>
|
||||
<span class='co'># [1] "Gram negative"</span>
|
||||
|
||||
<span class='fu'><a href='mo_property.html'>mo_ref</a></span>(<span class='st'>"E. coli"</span>)
|
||||
<span class='co'># [1] "Castellani and Chalmers, 1919"</span>
|
||||
|
||||
<span class='co'># Do not get mistaken - the package only includes microorganisms</span>
|
||||
<span class='fu'><a href='mo_property.html'>mo_phylum</a></span>(<span class='st'>"C. elegans"</span>)
|
||||
<span class='co'># [1] "Cyanobacteria" # Bacteria?!</span>
|
||||
<span class='fu'><a href='mo_property.html'>mo_fullname</a></span>(<span class='st'>"C. elegans"</span>)
|
||||
<span class='co'># [1] "Chroococcus limneticus elegans" # Because a microorganism was found</span>
|
||||
<span class='co'># }</span></pre>
|
||||
</div>
|
||||
<div class="col-md-3 hidden-xs hidden-sm" id="sidebar">
|
||||
<h2>Contents</h2>
|
||||
<ul class="nav nav-pills nav-stacked">
|
||||
|
||||
<li><a href="#itis">ITIS</a></li>
|
||||
|
||||
<li><a href="#read-more-on-our-website-">Read more on our website!</a></li>
|
||||
|
||||
<li><a href="#examples">Examples</a></li>
|
||||
</ul>
|
||||
|
||||
</div>
|
||||
</div>
|
||||
|
||||
<footer>
|
||||
<div class="copyright">
|
||||
<p>Developed by <a href='https://www.rug.nl/staff/m.s.berends/'>Matthijs S. Berends</a>, <a href='https://www.rug.nl/staff/c.f.luz/'>Christian F. Luz</a>, <a href='https://www.rug.nl/staff/c.glasner/'>Corinna Glasner</a>, <a href='https://www.rug.nl/staff/a.w.friedrich/'>Alex W. Friedrich</a>, <a href='https://www.rug.nl/staff/b.sinha/'>Bhanu N. M. Sinha</a>.</p>
|
||||
</div>
|
||||
|
||||
<div class="pkgdown">
|
||||
<p>Site built with <a href="https://pkgdown.r-lib.org/">pkgdown</a> 1.3.0.</p>
|
||||
</div>
|
||||
</footer>
|
||||
</div>
|
||||
|
||||
|
||||
<script src="https://cdnjs.cloudflare.com/ajax/libs/docsearch.js/2.6.1/docsearch.min.js" integrity="sha256-GKvGqXDznoRYHCwKXGnuchvKSwmx9SRMrZOTh2g4Sb0=" crossorigin="anonymous"></script>
|
||||
<script>
|
||||
docsearch({
|
||||
|
||||
|
||||
apiKey: 'f737050abfd4d726c63938e18f8c496e',
|
||||
indexName: 'amr',
|
||||
inputSelector: 'input#search-input.form-control',
|
||||
transformData: function(hits) {
|
||||
return hits.map(function (hit) {
|
||||
hit.url = updateHitURL(hit);
|
||||
return hit;
|
||||
});
|
||||
}
|
||||
});
|
||||
</script>
|
||||
|
||||
|
||||
</body>
|
||||
</html>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9026</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -306,10 +299,11 @@
|
||||
|
||||
<span class='co'># get frequencies of bacteria whose name start with 'Ent' or 'ent'</span>
|
||||
<span class='fu'><a href='https://www.rdocumentation.org/packages/base/topics/library'>library</a></span>(<span class='no'>dplyr</span>)
|
||||
<span class='fu'><a href='https://www.rdocumentation.org/packages/base/topics/library'>library</a></span>(<span class='no'>clean</span>)
|
||||
<span class='no'>septic_patients</span> <span class='kw'>%>%</span>
|
||||
<span class='fu'><a href='join.html'>left_join_microorganisms</a></span>() <span class='kw'>%>%</span>
|
||||
<span class='fu'><a href='https://dplyr.tidyverse.org/reference/filter.html'>filter</a></span>(<span class='no'>genus</span> <span class='kw'>%like%</span> <span class='st'>'^ent'</span>) <span class='kw'>%>%</span>
|
||||
<span class='fu'><a href='freq.html'>freq</a></span>(<span class='no'>genus</span>, <span class='no'>species</span>)
|
||||
<span class='fu'><a href='https://www.rdocumentation.org/packages/clean/topics/freq'>freq</a></span>(<span class='no'>genus</span>, <span class='no'>species</span>)
|
||||
<span class='co'># }</span></pre>
|
||||
</div>
|
||||
<div class="col-md-3 hidden-xs hidden-sm" id="sidebar">
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9007</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9026</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -244,7 +237,7 @@
|
||||
<pre class="usage"><span class='fu'>mdro</span>(<span class='no'>x</span>, <span class='kw'>guideline</span> <span class='kw'>=</span> <span class='kw'>NULL</span>, <span class='kw'>col_mo</span> <span class='kw'>=</span> <span class='kw'>NULL</span>, <span class='kw'>info</span> <span class='kw'>=</span> <span class='fl'>TRUE</span>,
|
||||
<span class='kw'>verbose</span> <span class='kw'>=</span> <span class='fl'>FALSE</span>, <span class='no'>...</span>)
|
||||
|
||||
<span class='fu'>brmo</span>(<span class='no'>...</span>, <span class='kw'>guideline</span> <span class='kw'>=</span> <span class='st'>"BRMO"</span>)
|
||||
<span class='fu'>brmo</span>(<span class='no'>x</span>, <span class='kw'>guideline</span> <span class='kw'>=</span> <span class='st'>"BRMO"</span>, <span class='no'>...</span>)
|
||||
|
||||
<span class='fu'>mrgn</span>(<span class='no'>x</span>, <span class='kw'>guideline</span> <span class='kw'>=</span> <span class='st'>"MRGN"</span>, <span class='no'>...</span>)
|
||||
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9026</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -245,7 +238,7 @@
|
||||
|
||||
<h2 class="hasAnchor" id="format"><a class="anchor" href="#format"></a>Format</h2>
|
||||
|
||||
<p>A <code><a href='https://www.rdocumentation.org/packages/base/topics/data.frame'>data.frame</a></code> with 67,906 observations and 16 variables:</p><dl class='dl-horizontal'>
|
||||
<p>A <code><a href='https://www.rdocumentation.org/packages/base/topics/data.frame'>data.frame</a></code> with 68,260 observations and 16 variables:</p><dl class='dl-horizontal'>
|
||||
<dt><code>mo</code></dt><dd><p>ID of microorganism as used by this package</p></dd>
|
||||
<dt><code>col_id</code></dt><dd><p>Catalogue of Life ID</p></dd>
|
||||
<dt><code>fullname</code></dt><dd><p>Full name, like <code>"Escherichia coli"</code></p></dd>
|
||||
@ -268,8 +261,8 @@
|
||||
<li><p>9 entries of <em>Streptococcus</em> (beta haemolytic groups A, B, C, D, F, G, H, K and unspecified)</p></li>
|
||||
<li><p>2 entries of <em>Staphylococcus</em> (coagulase-negative [CoNS] and coagulase-positive [CoPS])</p></li>
|
||||
<li><p>3 entries of Trichomonas (Trichomonas vaginalis, and its family and genus)</p></li>
|
||||
<li><p>3 other 'undefined' entries (unknown, unknown Gram negatives and unknown Gram positives)</p></li>
|
||||
<li><p>8,830 species from the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen) that are not in the Catalogue of Life</p></li>
|
||||
<li><p>5 other 'undefined' entries (unknown, unknown Gram negatives, unknown Gram positives, unknown yeast and unknown fungus)</p></li>
|
||||
<li><p>8,970 species from the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen) that are not in the Catalogue of Life</p></li>
|
||||
</ul>
|
||||
|
||||
<h2 class="hasAnchor" id="about-the-records-from-dsmz-see-source-"><a class="anchor" href="#about-the-records-from-dsmz-see-source-"></a>About the records from DSMZ (see source)</h2>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9026</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -245,7 +238,7 @@
|
||||
|
||||
<h2 class="hasAnchor" id="format"><a class="anchor" href="#format"></a>Format</h2>
|
||||
|
||||
<p>A <code><a href='https://www.rdocumentation.org/packages/base/topics/data.frame'>data.frame</a></code> with 21,342 observations and 4 variables:</p><dl class='dl-horizontal'>
|
||||
<p>A <code><a href='https://www.rdocumentation.org/packages/base/topics/data.frame'>data.frame</a></code> with 21,743 observations and 4 variables:</p><dl class='dl-horizontal'>
|
||||
<dt><code>col_id</code></dt><dd><p>Catalogue of Life ID that was originally given</p></dd>
|
||||
<dt><code>col_id_new</code></dt><dd><p>New Catalogue of Life ID that responds to an entry in the <code><a href='microorganisms.html'>microorganisms</a></code> data set</p></dd>
|
||||
<dt><code>fullname</code></dt><dd><p>Old full taxonomic name of the microorganism</p></dd>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9007</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -81,7 +81,7 @@ This is the fastest way to have your organisation (or analysis) specific codes p
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -159,13 +159,6 @@ This is the fastest way to have your organisation (or analysis) specific codes p
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -81,7 +81,7 @@ portion_R and portion_IR can be used to calculate resistance, portion_S and port
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9007</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -159,13 +159,6 @@ portion_R and portion_IR can be used to calculate resistance, portion_S and port
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9026</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
@ -291,11 +284,11 @@
|
||||
</tr>
|
||||
<tr>
|
||||
<th>model</th>
|
||||
<td><p>the statistical model of choice. Defaults to a generalised linear regression model with binomial distribution, assuming that a period of zero resistance was followed by a period of increasing resistance leading slowly to more and more resistance. See Details for valid options.</p></td>
|
||||
<td><p>the statistical model of choice. Defaults to a generalised linear regression model with binomial distribution (i.e. using <code><a href='https://www.rdocumentation.org/packages/stats/topics/glm'>glm</a>(..., family = <a href='https://www.rdocumentation.org/packages/stats/topics/family'>binomial</a>)</code>), assuming that a period of zero resistance was followed by a period of increasing resistance leading slowly to more and more resistance. See Details for valid options.</p></td>
|
||||
</tr>
|
||||
<tr>
|
||||
<th>I_as_S</th>
|
||||
<td><p>a logical to indicate whether values <code>I</code> should be treated as <code>S</code></p></td>
|
||||
<td><p>a logical to indicate whether values <code>I</code> should be treated as <code>S</code> (will otherwise be treated as <code>R</code>)</p></td>
|
||||
</tr>
|
||||
<tr>
|
||||
<th>preserve_measurements</th>
|
||||
|
||||
@ -80,7 +80,7 @@
|
||||
</button>
|
||||
<span class="navbar-brand">
|
||||
<a class="navbar-link" href="../index.html">AMR (for R)</a>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9005</span>
|
||||
<span class="version label label-default" data-toggle="tooltip" data-placement="bottom" title="Latest development version">0.7.1.9015</span>
|
||||
</span>
|
||||
</div>
|
||||
|
||||
@ -158,13 +158,6 @@
|
||||
Get properties of an antibiotic
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/freq.html">
|
||||
<span class="fa fa-sort-amount-down"></span>
|
||||
|
||||
Create frequency tables
|
||||
</a>
|
||||
</li>
|
||||
<li>
|
||||
<a href="../articles/benchmarks.html">
|
||||
<span class="fa fa-shipping-fast"></span>
|
||||
|
||||