Built site for AMR@2.1.1.9133: 9520977
2
404.html
@ -30,7 +30,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="https://msberends.github.io/AMR/index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -29,7 +29,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -88,7 +88,7 @@
|
||||
website update since they are based on randomly created values and the
|
||||
page was written in <a href="https://rmarkdown.rstudio.com/" class="external-link">R
|
||||
Markdown</a>. However, the methodology remains unchanged. This page was
|
||||
generated on 17 January 2025.</p>
|
||||
generated on 27 January 2025.</p>
|
||||
<div class="section level2">
|
||||
<h2 id="introduction">Introduction<a class="anchor" aria-label="anchor" href="#introduction"></a>
|
||||
</h2>
|
||||
@ -144,21 +144,21 @@ make the structure of your data generally look like this:</p>
|
||||
</tr></thead>
|
||||
<tbody>
|
||||
<tr class="odd">
|
||||
<td align="center">2025-01-17</td>
|
||||
<td align="center">2025-01-27</td>
|
||||
<td align="center">abcd</td>
|
||||
<td align="center">Escherichia coli</td>
|
||||
<td align="center">S</td>
|
||||
<td align="center">S</td>
|
||||
</tr>
|
||||
<tr class="even">
|
||||
<td align="center">2025-01-17</td>
|
||||
<td align="center">2025-01-27</td>
|
||||
<td align="center">abcd</td>
|
||||
<td align="center">Escherichia coli</td>
|
||||
<td align="center">S</td>
|
||||
<td align="center">R</td>
|
||||
</tr>
|
||||
<tr class="odd">
|
||||
<td align="center">2025-01-17</td>
|
||||
<td align="center">2025-01-27</td>
|
||||
<td align="center">efgh</td>
|
||||
<td align="center">Escherichia coli</td>
|
||||
<td align="center">R</td>
|
||||
@ -691,26 +691,9 @@ previously mentioned antibiotic class selectors:</p>
|
||||
<div class="sourceCode" id="cb16"><pre class="downlit sourceCode r">
|
||||
<code class="sourceCode R"><span><span class="fu"><a href="../reference/antibiogram.html">antibiogram</a></span><span class="op">(</span><span class="va">example_isolates</span>,</span>
|
||||
<span> antibiotics <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="fu"><a href="../reference/antimicrobial_class_selectors.html">aminoglycosides</a></span><span class="op">(</span><span class="op">)</span>, <span class="fu"><a href="../reference/antimicrobial_class_selectors.html">carbapenems</a></span><span class="op">(</span><span class="op">)</span><span class="op">)</span><span class="op">)</span></span>
|
||||
<span><span class="co">#> ℹ The function aminoglycosides() should be used inside a dplyr verb or</span></span>
|
||||
<span><span class="co">#> data.frame call, e.g.:</span></span>
|
||||
<span><span class="co">#> • your_data %>% select(aminoglycosides())</span></span>
|
||||
<span><span class="co">#> • your_data %>% select(column_a, column_b, aminoglycosides())</span></span>
|
||||
<span><span class="co">#> • your_data %>% filter(any(aminoglycosides() == "R"))</span></span>
|
||||
<span><span class="co">#> • your_data[, aminoglycosides()]</span></span>
|
||||
<span><span class="co">#> • your_data[, c("column_a", "column_b", aminoglycosides())]</span></span>
|
||||
<span><span class="co">#> </span></span>
|
||||
<span><span class="co">#> Now returning a vector of all possible antimicrobials that</span></span>
|
||||
<span><span class="co">#> aminoglycosides() can select.</span></span>
|
||||
<span><span class="co">#> ℹ The function carbapenems() should be used inside a dplyr verb or</span></span>
|
||||
<span><span class="co">#> data.frame call, e.g.:</span></span>
|
||||
<span><span class="co">#> • your_data %>% select(carbapenems())</span></span>
|
||||
<span><span class="co">#> • your_data %>% select(column_a, column_b, carbapenems())</span></span>
|
||||
<span><span class="co">#> • your_data %>% filter(any(carbapenems() == "R"))</span></span>
|
||||
<span><span class="co">#> • your_data[, carbapenems()]</span></span>
|
||||
<span><span class="co">#> • your_data[, c("column_a", "column_b", carbapenems())]</span></span>
|
||||
<span><span class="co">#> </span></span>
|
||||
<span><span class="co">#> Now returning a vector of all possible antimicrobials that carbapenems()</span></span>
|
||||
<span><span class="co">#> can select.</span></span></code></pre></div>
|
||||
<span><span class="co">#> ℹ For aminoglycosides() using columns 'GEN' (gentamicin), 'TOB'</span></span>
|
||||
<span><span class="co">#> (tobramycin), 'AMK' (amikacin), and 'KAN' (kanamycin)</span></span>
|
||||
<span><span class="co">#> ℹ For carbapenems() using columns 'IPM' (imipenem) and 'MEM' (meropenem)</span></span></code></pre></div>
|
||||
<table class="table">
|
||||
<colgroup>
|
||||
<col width="16%">
|
||||
@ -838,16 +821,8 @@ language to be Spanish using the <code>language</code> argument:</p>
|
||||
<span> antibiotics <span class="op">=</span> <span class="fu"><a href="../reference/antimicrobial_class_selectors.html">aminoglycosides</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> ab_transform <span class="op">=</span> <span class="st">"name"</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="st">"es"</span><span class="op">)</span></span>
|
||||
<span><span class="co">#> ℹ The function aminoglycosides() should be used inside a dplyr verb or</span></span>
|
||||
<span><span class="co">#> data.frame call, e.g.:</span></span>
|
||||
<span><span class="co">#> • your_data %>% select(aminoglycosides())</span></span>
|
||||
<span><span class="co">#> • your_data %>% select(column_a, column_b, aminoglycosides())</span></span>
|
||||
<span><span class="co">#> • your_data %>% filter(any(aminoglycosides() == "R"))</span></span>
|
||||
<span><span class="co">#> • your_data[, aminoglycosides()]</span></span>
|
||||
<span><span class="co">#> • your_data[, c("column_a", "column_b", aminoglycosides())]</span></span>
|
||||
<span><span class="co">#> </span></span>
|
||||
<span><span class="co">#> Now returning a vector of all possible antimicrobials that</span></span>
|
||||
<span><span class="co">#> aminoglycosides() can select.</span></span></code></pre></div>
|
||||
<span><span class="co">#> ℹ For aminoglycosides() using columns 'GEN' (gentamicin), 'TOB'</span></span>
|
||||
<span><span class="co">#> (tobramycin), 'AMK' (amikacin), and 'KAN' (kanamycin)</span></span></code></pre></div>
|
||||
<table class="table">
|
||||
<colgroup>
|
||||
<col width="20%">
|
||||
@ -970,26 +945,9 @@ argument must be used. This can be any column in the data, or e.g. an
|
||||
<code class="sourceCode R"><span><span class="fu"><a href="../reference/antibiogram.html">antibiogram</a></span><span class="op">(</span><span class="va">example_isolates</span>,</span>
|
||||
<span> antibiotics <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="fu"><a href="../reference/antimicrobial_class_selectors.html">aminoglycosides</a></span><span class="op">(</span><span class="op">)</span>, <span class="fu"><a href="../reference/antimicrobial_class_selectors.html">carbapenems</a></span><span class="op">(</span><span class="op">)</span><span class="op">)</span>,</span>
|
||||
<span> syndromic_group <span class="op">=</span> <span class="st">"ward"</span><span class="op">)</span></span>
|
||||
<span><span class="co">#> ℹ The function aminoglycosides() should be used inside a dplyr verb or</span></span>
|
||||
<span><span class="co">#> data.frame call, e.g.:</span></span>
|
||||
<span><span class="co">#> • your_data %>% select(aminoglycosides())</span></span>
|
||||
<span><span class="co">#> • your_data %>% select(column_a, column_b, aminoglycosides())</span></span>
|
||||
<span><span class="co">#> • your_data %>% filter(any(aminoglycosides() == "R"))</span></span>
|
||||
<span><span class="co">#> • your_data[, aminoglycosides()]</span></span>
|
||||
<span><span class="co">#> • your_data[, c("column_a", "column_b", aminoglycosides())]</span></span>
|
||||
<span><span class="co">#> </span></span>
|
||||
<span><span class="co">#> Now returning a vector of all possible antimicrobials that</span></span>
|
||||
<span><span class="co">#> aminoglycosides() can select.</span></span>
|
||||
<span><span class="co">#> ℹ The function carbapenems() should be used inside a dplyr verb or</span></span>
|
||||
<span><span class="co">#> data.frame call, e.g.:</span></span>
|
||||
<span><span class="co">#> • your_data %>% select(carbapenems())</span></span>
|
||||
<span><span class="co">#> • your_data %>% select(column_a, column_b, carbapenems())</span></span>
|
||||
<span><span class="co">#> • your_data %>% filter(any(carbapenems() == "R"))</span></span>
|
||||
<span><span class="co">#> • your_data[, carbapenems()]</span></span>
|
||||
<span><span class="co">#> • your_data[, c("column_a", "column_b", carbapenems())]</span></span>
|
||||
<span><span class="co">#> </span></span>
|
||||
<span><span class="co">#> Now returning a vector of all possible antimicrobials that carbapenems()</span></span>
|
||||
<span><span class="co">#> can select.</span></span></code></pre></div>
|
||||
<span><span class="co">#> ℹ For aminoglycosides() using columns 'GEN' (gentamicin), 'TOB'</span></span>
|
||||
<span><span class="co">#> (tobramycin), 'AMK' (amikacin), and 'KAN' (kanamycin)</span></span>
|
||||
<span><span class="co">#> ℹ For carbapenems() using columns 'IPM' (imipenem) and 'MEM' (meropenem)</span></span></code></pre></div>
|
||||
<table class="table">
|
||||
<colgroup>
|
||||
<col width="13%">
|
||||
|
||||
@ -29,7 +29,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -200,6 +200,129 @@ the standard name, “Ciprofloxacin”.</p></li>
|
||||
</div>
|
||||
</div>
|
||||
<div class="section level3">
|
||||
<h3 id="calculating-amr">Calculating AMR<a class="anchor" aria-label="anchor" href="#calculating-amr"></a>
|
||||
</h3>
|
||||
<div class="sourceCode" id="cb5"><pre class="sourceCode python"><code class="sourceCode python"><span id="cb5-1"><a href="#cb5-1" tabindex="-1"></a><span class="im">import</span> AMR</span>
|
||||
<span id="cb5-2"><a href="#cb5-2" tabindex="-1"></a><span class="im">import</span> pandas <span class="im">as</span> pd</span>
|
||||
<span id="cb5-3"><a href="#cb5-3" tabindex="-1"></a></span>
|
||||
<span id="cb5-4"><a href="#cb5-4" tabindex="-1"></a>df <span class="op">=</span> AMR.example_isolates</span>
|
||||
<span id="cb5-5"><a href="#cb5-5" tabindex="-1"></a>result <span class="op">=</span> AMR.resistance(df[<span class="st">"AMX"</span>])</span>
|
||||
<span id="cb5-6"><a href="#cb5-6" tabindex="-1"></a><span class="bu">print</span>(result)</span></code></pre></div>
|
||||
<pre><code>[0.59555556]</code></pre>
|
||||
</div>
|
||||
<div class="section level3">
|
||||
<h3 id="generating-antibiograms">Generating Antibiograms<a class="anchor" aria-label="anchor" href="#generating-antibiograms"></a>
|
||||
</h3>
|
||||
<p>One of the core functions of the <code>AMR</code> package is
|
||||
generating an antibiogram, a table that summarises the antimicrobial
|
||||
susceptibility of bacterial isolates. Here’s how you can generate an
|
||||
antibiogram from Python:</p>
|
||||
<div class="sourceCode" id="cb7"><pre class="sourceCode python"><code class="sourceCode python"><span id="cb7-1"><a href="#cb7-1" tabindex="-1"></a>result2a <span class="op">=</span> AMR.antibiogram(df[[<span class="st">"mo"</span>, <span class="st">"AMX"</span>, <span class="st">"CIP"</span>, <span class="st">"TZP"</span>]])</span>
|
||||
<span id="cb7-2"><a href="#cb7-2" tabindex="-1"></a><span class="bu">print</span>(result2a)</span></code></pre></div>
|
||||
<table class="table">
|
||||
<colgroup>
|
||||
<col width="22%">
|
||||
<col width="22%">
|
||||
<col width="22%">
|
||||
<col width="33%">
|
||||
</colgroup>
|
||||
<thead><tr class="header">
|
||||
<th>Pathogen</th>
|
||||
<th>Amoxicillin</th>
|
||||
<th>Ciprofloxacin</th>
|
||||
<th>Piperacillin/tazobactam</th>
|
||||
</tr></thead>
|
||||
<tbody>
|
||||
<tr class="odd">
|
||||
<td>CoNS</td>
|
||||
<td>7% (10/142)</td>
|
||||
<td>73% (183/252)</td>
|
||||
<td>30% (10/33)</td>
|
||||
</tr>
|
||||
<tr class="even">
|
||||
<td>E. coli</td>
|
||||
<td>50% (196/392)</td>
|
||||
<td>88% (399/456)</td>
|
||||
<td>94% (393/416)</td>
|
||||
</tr>
|
||||
<tr class="odd">
|
||||
<td>K. pneumoniae</td>
|
||||
<td>0% (0/58)</td>
|
||||
<td>96% (53/55)</td>
|
||||
<td>89% (47/53)</td>
|
||||
</tr>
|
||||
<tr class="even">
|
||||
<td>P. aeruginosa</td>
|
||||
<td>0% (0/30)</td>
|
||||
<td>100% (30/30)</td>
|
||||
<td>None</td>
|
||||
</tr>
|
||||
<tr class="odd">
|
||||
<td>P. mirabilis</td>
|
||||
<td>None</td>
|
||||
<td>94% (34/36)</td>
|
||||
<td>None</td>
|
||||
</tr>
|
||||
<tr class="even">
|
||||
<td>S. aureus</td>
|
||||
<td>6% (8/131)</td>
|
||||
<td>90% (171/191)</td>
|
||||
<td>None</td>
|
||||
</tr>
|
||||
<tr class="odd">
|
||||
<td>S. epidermidis</td>
|
||||
<td>1% (1/91)</td>
|
||||
<td>64% (87/136)</td>
|
||||
<td>None</td>
|
||||
</tr>
|
||||
<tr class="even">
|
||||
<td>S. hominis</td>
|
||||
<td>None</td>
|
||||
<td>80% (56/70)</td>
|
||||
<td>None</td>
|
||||
</tr>
|
||||
<tr class="odd">
|
||||
<td>S. pneumoniae</td>
|
||||
<td>100% (112/112)</td>
|
||||
<td>None</td>
|
||||
<td>100% (112/112)</td>
|
||||
</tr>
|
||||
</tbody>
|
||||
</table>
|
||||
<div class="sourceCode" id="cb8"><pre class="sourceCode python"><code class="sourceCode python"><span id="cb8-1"><a href="#cb8-1" tabindex="-1"></a>result2b <span class="op">=</span> AMR.antibiogram(df[[<span class="st">"mo"</span>, <span class="st">"AMX"</span>, <span class="st">"CIP"</span>, <span class="st">"TZP"</span>]], mo_transform <span class="op">=</span> <span class="st">"gramstain"</span>)</span>
|
||||
<span id="cb8-2"><a href="#cb8-2" tabindex="-1"></a><span class="bu">print</span>(result2b)</span></code></pre></div>
|
||||
<table class="table">
|
||||
<colgroup>
|
||||
<col width="20%">
|
||||
<col width="22%">
|
||||
<col width="23%">
|
||||
<col width="33%">
|
||||
</colgroup>
|
||||
<thead><tr class="header">
|
||||
<th>Pathogen</th>
|
||||
<th>Amoxicillin</th>
|
||||
<th>Ciprofloxacin</th>
|
||||
<th>Piperacillin/tazobactam</th>
|
||||
</tr></thead>
|
||||
<tbody>
|
||||
<tr class="odd">
|
||||
<td>Gram-negative</td>
|
||||
<td>36% (226/631)</td>
|
||||
<td>91% (621/684)</td>
|
||||
<td>88% (565/641)</td>
|
||||
</tr>
|
||||
<tr class="even">
|
||||
<td>Gram-positive</td>
|
||||
<td>43% (305/703)</td>
|
||||
<td>77% (560/724)</td>
|
||||
<td>86% (296/345)</td>
|
||||
</tr>
|
||||
</tbody>
|
||||
</table>
|
||||
<p>In this example, we generate an antibiogram by selecting various
|
||||
antibiotics.</p>
|
||||
</div>
|
||||
<div class="section level3">
|
||||
<h3 id="taxonomic-data-sets-now-in-python">Taxonomic Data Sets Now in Python!<a class="anchor" aria-label="anchor" href="#taxonomic-data-sets-now-in-python"></a>
|
||||
</h3>
|
||||
<p>As a Python user, you might like that the most important data sets of
|
||||
@ -207,7 +330,7 @@ the <code>AMR</code> R package, <code>microorganisms</code>,
|
||||
<code>antibiotics</code>, <code>clinical_breakpoints</code>, and
|
||||
<code>example_isolates</code>, are now available as regular Python data
|
||||
frames:</p>
|
||||
<div class="sourceCode" id="cb5"><pre class="sourceCode python"><code class="sourceCode python"><span id="cb5-1"><a href="#cb5-1" tabindex="-1"></a>AMR.microorganisms</span></code></pre></div>
|
||||
<div class="sourceCode" id="cb9"><pre class="sourceCode python"><code class="sourceCode python"><span id="cb9-1"><a href="#cb9-1" tabindex="-1"></a>AMR.microorganisms</span></code></pre></div>
|
||||
<table class="table">
|
||||
<colgroup>
|
||||
<col width="11%">
|
||||
@ -342,7 +465,7 @@ frames:</p>
|
||||
</tr>
|
||||
</tbody>
|
||||
</table>
|
||||
<div class="sourceCode" id="cb6"><pre class="sourceCode python"><code class="sourceCode python"><span id="cb6-1"><a href="#cb6-1" tabindex="-1"></a>AMR.antibiotics</span></code></pre></div>
|
||||
<div class="sourceCode" id="cb10"><pre class="sourceCode python"><code class="sourceCode python"><span id="cb10-1"><a href="#cb10-1" tabindex="-1"></a>AMR.antibiotics</span></code></pre></div>
|
||||
<table style="width:100%;" class="table">
|
||||
<colgroup>
|
||||
<col width="4%">
|
||||
@ -478,129 +601,6 @@ frames:</p>
|
||||
</tbody>
|
||||
</table>
|
||||
</div>
|
||||
<div class="section level3">
|
||||
<h3 id="calculating-amr">Calculating AMR<a class="anchor" aria-label="anchor" href="#calculating-amr"></a>
|
||||
</h3>
|
||||
<div class="sourceCode" id="cb7"><pre class="sourceCode python"><code class="sourceCode python"><span id="cb7-1"><a href="#cb7-1" tabindex="-1"></a><span class="im">import</span> AMR</span>
|
||||
<span id="cb7-2"><a href="#cb7-2" tabindex="-1"></a><span class="im">import</span> pandas <span class="im">as</span> pd</span>
|
||||
<span id="cb7-3"><a href="#cb7-3" tabindex="-1"></a></span>
|
||||
<span id="cb7-4"><a href="#cb7-4" tabindex="-1"></a>df <span class="op">=</span> AMR.example_isolates</span>
|
||||
<span id="cb7-5"><a href="#cb7-5" tabindex="-1"></a>result <span class="op">=</span> AMR.resistance(df[<span class="st">"AMX"</span>])</span>
|
||||
<span id="cb7-6"><a href="#cb7-6" tabindex="-1"></a><span class="bu">print</span>(result)</span></code></pre></div>
|
||||
<pre><code>[0.59555556]</code></pre>
|
||||
</div>
|
||||
<div class="section level3">
|
||||
<h3 id="generating-antibiograms">Generating Antibiograms<a class="anchor" aria-label="anchor" href="#generating-antibiograms"></a>
|
||||
</h3>
|
||||
<p>One of the core functions of the <code>AMR</code> package is
|
||||
generating an antibiogram, a table that summarises the antimicrobial
|
||||
susceptibility of bacterial isolates. Here’s how you can generate an
|
||||
antibiogram from Python:</p>
|
||||
<div class="sourceCode" id="cb9"><pre class="sourceCode python"><code class="sourceCode python"><span id="cb9-1"><a href="#cb9-1" tabindex="-1"></a>result2a <span class="op">=</span> AMR.antibiogram(df[[<span class="st">"mo"</span>, <span class="st">"AMX"</span>, <span class="st">"CIP"</span>, <span class="st">"TZP"</span>]])</span>
|
||||
<span id="cb9-2"><a href="#cb9-2" tabindex="-1"></a><span class="bu">print</span>(result2a)</span></code></pre></div>
|
||||
<table class="table">
|
||||
<colgroup>
|
||||
<col width="22%">
|
||||
<col width="22%">
|
||||
<col width="22%">
|
||||
<col width="33%">
|
||||
</colgroup>
|
||||
<thead><tr class="header">
|
||||
<th>Pathogen</th>
|
||||
<th>Amoxicillin</th>
|
||||
<th>Ciprofloxacin</th>
|
||||
<th>Piperacillin/tazobactam</th>
|
||||
</tr></thead>
|
||||
<tbody>
|
||||
<tr class="odd">
|
||||
<td>CoNS</td>
|
||||
<td>7% (10/142)</td>
|
||||
<td>73% (183/252)</td>
|
||||
<td>30% (10/33)</td>
|
||||
</tr>
|
||||
<tr class="even">
|
||||
<td>E. coli</td>
|
||||
<td>50% (196/392)</td>
|
||||
<td>88% (399/456)</td>
|
||||
<td>94% (393/416)</td>
|
||||
</tr>
|
||||
<tr class="odd">
|
||||
<td>K. pneumoniae</td>
|
||||
<td>0% (0/58)</td>
|
||||
<td>96% (53/55)</td>
|
||||
<td>89% (47/53)</td>
|
||||
</tr>
|
||||
<tr class="even">
|
||||
<td>P. aeruginosa</td>
|
||||
<td>0% (0/30)</td>
|
||||
<td>100% (30/30)</td>
|
||||
<td>None</td>
|
||||
</tr>
|
||||
<tr class="odd">
|
||||
<td>P. mirabilis</td>
|
||||
<td>None</td>
|
||||
<td>94% (34/36)</td>
|
||||
<td>None</td>
|
||||
</tr>
|
||||
<tr class="even">
|
||||
<td>S. aureus</td>
|
||||
<td>6% (8/131)</td>
|
||||
<td>90% (171/191)</td>
|
||||
<td>None</td>
|
||||
</tr>
|
||||
<tr class="odd">
|
||||
<td>S. epidermidis</td>
|
||||
<td>1% (1/91)</td>
|
||||
<td>64% (87/136)</td>
|
||||
<td>None</td>
|
||||
</tr>
|
||||
<tr class="even">
|
||||
<td>S. hominis</td>
|
||||
<td>None</td>
|
||||
<td>80% (56/70)</td>
|
||||
<td>None</td>
|
||||
</tr>
|
||||
<tr class="odd">
|
||||
<td>S. pneumoniae</td>
|
||||
<td>100% (112/112)</td>
|
||||
<td>None</td>
|
||||
<td>100% (112/112)</td>
|
||||
</tr>
|
||||
</tbody>
|
||||
</table>
|
||||
<div class="sourceCode" id="cb10"><pre class="sourceCode python"><code class="sourceCode python"><span id="cb10-1"><a href="#cb10-1" tabindex="-1"></a>result2b <span class="op">=</span> AMR.antibiogram(df[[<span class="st">"mo"</span>, <span class="st">"AMX"</span>, <span class="st">"CIP"</span>, <span class="st">"TZP"</span>]], mo_transform <span class="op">=</span> <span class="st">"gramstain"</span>)</span>
|
||||
<span id="cb10-2"><a href="#cb10-2" tabindex="-1"></a><span class="bu">print</span>(result2b)</span></code></pre></div>
|
||||
<table class="table">
|
||||
<colgroup>
|
||||
<col width="20%">
|
||||
<col width="22%">
|
||||
<col width="23%">
|
||||
<col width="33%">
|
||||
</colgroup>
|
||||
<thead><tr class="header">
|
||||
<th>Pathogen</th>
|
||||
<th>Amoxicillin</th>
|
||||
<th>Ciprofloxacin</th>
|
||||
<th>Piperacillin/tazobactam</th>
|
||||
</tr></thead>
|
||||
<tbody>
|
||||
<tr class="odd">
|
||||
<td>Gram-negative</td>
|
||||
<td>36% (226/631)</td>
|
||||
<td>91% (621/684)</td>
|
||||
<td>88% (565/641)</td>
|
||||
</tr>
|
||||
<tr class="even">
|
||||
<td>Gram-positive</td>
|
||||
<td>43% (305/703)</td>
|
||||
<td>77% (560/724)</td>
|
||||
<td>86% (296/345)</td>
|
||||
</tr>
|
||||
</tbody>
|
||||
</table>
|
||||
<p>In this example, we generate an antibiogram by selecting various
|
||||
antibiotics.</p>
|
||||
</div>
|
||||
</div>
|
||||
<div class="section level2">
|
||||
<h2 id="conclusion">Conclusion<a class="anchor" aria-label="anchor" href="#conclusion"></a>
|
||||
|
||||
@ -29,7 +29,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -134,13 +134,13 @@ package.</p>
|
||||
<span><span class="co">#> <span style="color: #00BB00;">✔</span> <span style="color: #0000BB;">infer </span> 1.0.7 <span style="color: #00BB00;">✔</span> <span style="color: #0000BB;">tune </span> 1.2.1</span></span>
|
||||
<span><span class="co">#> <span style="color: #00BB00;">✔</span> <span style="color: #0000BB;">modeldata </span> 1.4.0 <span style="color: #00BB00;">✔</span> <span style="color: #0000BB;">workflows </span> 1.1.4</span></span>
|
||||
<span><span class="co">#> <span style="color: #00BB00;">✔</span> <span style="color: #0000BB;">parsnip </span> 1.2.1 <span style="color: #00BB00;">✔</span> <span style="color: #0000BB;">workflowsets</span> 1.1.0</span></span>
|
||||
<span><span class="co">#> <span style="color: #00BB00;">✔</span> <span style="color: #0000BB;">purrr </span> 1.0.2 <span style="color: #00BB00;">✔</span> <span style="color: #0000BB;">yardstick </span> 1.3.1</span></span>
|
||||
<span><span class="co">#> <span style="color: #00BB00;">✔</span> <span style="color: #0000BB;">purrr </span> 1.0.2 <span style="color: #00BB00;">✔</span> <span style="color: #0000BB;">yardstick </span> 1.3.2</span></span>
|
||||
<span><span class="co">#> ── <span style="font-weight: bold;">Conflicts</span> ───────────────────────────────────────── tidymodels_conflicts() ──</span></span>
|
||||
<span><span class="co">#> <span style="color: #BB0000;">✖</span> <span style="color: #0000BB;">purrr</span>::<span style="color: #00BB00;">discard()</span> masks <span style="color: #0000BB;">scales</span>::discard()</span></span>
|
||||
<span><span class="co">#> <span style="color: #BB0000;">✖</span> <span style="color: #0000BB;">dplyr</span>::<span style="color: #00BB00;">filter()</span> masks <span style="color: #0000BB;">stats</span>::filter()</span></span>
|
||||
<span><span class="co">#> <span style="color: #BB0000;">✖</span> <span style="color: #0000BB;">dplyr</span>::<span style="color: #00BB00;">lag()</span> masks <span style="color: #0000BB;">stats</span>::lag()</span></span>
|
||||
<span><span class="co">#> <span style="color: #BB0000;">✖</span> <span style="color: #0000BB;">recipes</span>::<span style="color: #00BB00;">step()</span> masks <span style="color: #0000BB;">stats</span>::step()</span></span>
|
||||
<span><span class="co">#> <span style="color: #0000BB;">•</span> Dig deeper into tidy modeling with R at <span style="color: #00BB00;">https://www.tmwr.org</span></span></span>
|
||||
<span><span class="co">#> <span style="color: #0000BB;">•</span> Use <span style="color: #00BB00;">tidymodels_prefer()</span> to resolve common conflicts.</span></span>
|
||||
<span><span class="kw"><a href="https://rdrr.io/r/base/library.html" class="external-link">library</a></span><span class="op">(</span><span class="va"><a href="https://msberends.github.io/AMR/">AMR</a></span><span class="op">)</span> <span class="co"># For AMR data analysis</span></span>
|
||||
<span></span>
|
||||
<span><span class="co"># Load the example_isolates dataset</span></span>
|
||||
|
||||
@ -29,7 +29,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -29,7 +29,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -29,7 +29,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -29,7 +29,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -29,7 +29,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -77,7 +77,7 @@
|
||||
<main id="main" class="col-md-9"><div class="page-header">
|
||||
<img src="../logo.svg" class="logo" alt=""><h1>Data sets for download / own use</h1>
|
||||
|
||||
<h4 data-toc-skip class="date">17 January 2025</h4>
|
||||
<h4 data-toc-skip class="date">27 January 2025</h4>
|
||||
|
||||
<small class="dont-index">Source: <a href="https://github.com/msberends/AMR/blob/main/vignettes/datasets.Rmd" class="external-link"><code>vignettes/datasets.Rmd</code></a></small>
|
||||
<div class="d-none name"><code>datasets.Rmd</code></div>
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -29,7 +29,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -29,7 +29,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -32,7 +32,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -680,7 +680,7 @@
|
||||
<p>Manually, using:</p>
|
||||
<div class="sourceCode" id="cb12"><pre class="downlit sourceCode r">
|
||||
<code class="sourceCode R"><span><span class="fu"><a href="https://rdrr.io/r/utils/install.packages.html" class="external-link">install.packages</a></span><span class="op">(</span><span class="st">"remotes"</span><span class="op">)</span> <span class="co"># if you haven't already</span></span>
|
||||
<span><span class="fu">remotes</span><span class="fu">::</span><span class="fu">install_github</span><span class="op">(</span><span class="st">"msberends/AMR"</span><span class="op">)</span></span></code></pre></div>
|
||||
<span><span class="fu">remotes</span><span class="fu">::</span><span class="fu"><a href="https://remotes.r-lib.org/reference/install_github.html" class="external-link">install_github</a></span><span class="op">(</span><span class="st">"msberends/AMR"</span><span class="op">)</span></span></code></pre></div>
|
||||
</li>
|
||||
<li>
|
||||
<p>Automatically, using the <a href="https://ropensci.org/r-universe/" class="external-link">rOpenSci R-universe platform</a>, by adding <a href="https://msberends.r-universe.dev" class="external-link">our R-universe address</a> to your list of repositories (‘repos’):</p>
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -48,18 +48,18 @@
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
<h2 class="pkg-version" data-toc-text="2.1.1.9125" id="amr-2119125">AMR 2.1.1.9125<a class="anchor" aria-label="anchor" href="#amr-2119125"></a></h2>
|
||||
<h2 class="pkg-version" data-toc-text="2.1.1.9133" id="amr-2119133">AMR 2.1.1.9133<a class="anchor" aria-label="anchor" href="#amr-2119133"></a></h2>
|
||||
<p><em>(this beta version will eventually become v3.0. We’re happy to reach a new major milestone soon, which will be all about the new One Health support! Install this beta using <a href="https://msberends.github.io/AMR/#latest-development-version">the instructions here</a>.)</em></p>
|
||||
<div class="section level5">
|
||||
<h5 id="a-new-milestone-amr-v30-with-one-health-support--human--veterinary--environmental-2-1-1-9125">A New Milestone: AMR v3.0 with One Health Support (= Human + Veterinary + Environmental)<a class="anchor" aria-label="anchor" href="#a-new-milestone-amr-v30-with-one-health-support--human--veterinary--environmental-2-1-1-9125"></a></h5>
|
||||
<h5 id="a-new-milestone-amr-v30-with-one-health-support--human--veterinary--environmental-2-1-1-9133">A New Milestone: AMR v3.0 with One Health Support (= Human + Veterinary + Environmental)<a class="anchor" aria-label="anchor" href="#a-new-milestone-amr-v30-with-one-health-support--human--veterinary--environmental-2-1-1-9133"></a></h5>
|
||||
<p>This package now supports not only tools for AMR data analysis in clinical settings, but also for veterinary and environmental microbiology. This was made possible through a collaboration with the <a href="https://www.upei.ca/avc" class="external-link">University of Prince Edward Island’s Atlantic Veterinary College</a>, Canada. To celebrate this great improvement of the package, we also updated the package logo to reflect this change.</p>
|
||||
</div>
|
||||
<div class="section level3">
|
||||
<h3 id="breaking-2-1-1-9125">Breaking<a class="anchor" aria-label="anchor" href="#breaking-2-1-1-9125"></a></h3>
|
||||
<ul><li>Removed all functions and references that used the deprecated <code>rsi</code> class, which were all replaced with their <code>sir</code> equivalents over a year ago</li>
|
||||
<h3 id="breaking-2-1-1-9133">Breaking<a class="anchor" aria-label="anchor" href="#breaking-2-1-1-9133"></a></h3>
|
||||
<ul><li>Removed all functions and references that used the deprecated <code>rsi</code> class, which were all replaced with their <code>sir</code> equivalents two years ago</li>
|
||||
</ul></div>
|
||||
<div class="section level3">
|
||||
<h3 id="new-2-1-1-9125">New<a class="anchor" aria-label="anchor" href="#new-2-1-1-9125"></a></h3>
|
||||
<h3 id="new-2-1-1-9133">New<a class="anchor" aria-label="anchor" href="#new-2-1-1-9133"></a></h3>
|
||||
<ul><li>
|
||||
<strong>One Health implementation</strong>
|
||||
<ul><li>Function <code><a href="../reference/as.sir.html">as.sir()</a></code> now has extensive support for veterinary breakpoints from CLSI. Use <code>breakpoint_type = "animal"</code> and set the <code>host</code> argument to a variable that contains animal species names.</li>
|
||||
@ -72,6 +72,10 @@
|
||||
<code><a href="../reference/ab_property.html">ab_url()</a></code> now supports retrieving the WHOCC url of their ATCvet pages</li>
|
||||
</ul></li>
|
||||
<li>
|
||||
<strong>Support for WISCA antibiograms</strong>
|
||||
<ul><li>The <code><a href="../reference/antibiogram.html">antibiogram()</a></code> function now supports creating true Weighted-Incidence Syndromic Combination Antibiograms (WISCA), a powerful Bayesian method for estimating regimen coverage probabilities using pathogen incidence and antimicrobial susceptibility data. WISCA offers improved precision for syndrome-specific treatment, even in datasets with sparse data. A dedicated <code><a href="../reference/antibiogram.html">wisca()</a></code> function is also available for easy usage.</li>
|
||||
</ul></li>
|
||||
<li>
|
||||
<strong>Major update to fungal taxonomy and tools for mycologists</strong>
|
||||
<ul><li>MycoBank has now been integrated as the primary taxonomic source for fungi. The <code>microorganisms</code> data set has been enriched with new columns (<code>mycobank</code>, <code>mycobank_parent</code>, and <code>mycobank_renamed_to</code>) that provide detailed information for fungal species.</li>
|
||||
<li>A remarkable addition of over 20,000 new fungal records</li>
|
||||
@ -100,11 +104,12 @@
|
||||
</ul></li>
|
||||
<li>
|
||||
<strong>Other</strong>
|
||||
<ul><li>New function <code><a href="../reference/mo_property.html">mo_group_members()</a></code> to retrieve the member microorganisms of a microorganism group. For example, <code>mo_group_members("Strep group C")</code> returns a vector of all microorganisms that belong to that group.</li>
|
||||
<ul><li>New function <code><a href="../reference/top_n_microorganisms.html">top_n_microorganisms()</a></code> to filter a data set to the top <em>n</em> of any taxonomic property, e.g., filter to the top 3 species, filter to any species in the top 5 genera, or filter to the top 3 species in each of the top 5 genera</li>
|
||||
<li>New function <code><a href="../reference/mo_property.html">mo_group_members()</a></code> to retrieve the member microorganisms of a microorganism group. For example, <code>mo_group_members("Strep group C")</code> returns a vector of all microorganisms that belong to that group.</li>
|
||||
</ul></li>
|
||||
</ul></div>
|
||||
<div class="section level3">
|
||||
<h3 id="changed-2-1-1-9125">Changed<a class="anchor" aria-label="anchor" href="#changed-2-1-1-9125"></a></h3>
|
||||
<h3 id="changed-2-1-1-9133">Changed<a class="anchor" aria-label="anchor" href="#changed-2-1-1-9133"></a></h3>
|
||||
<ul><li>SIR interpretation
|
||||
<ul><li>It is now possible to use column names for argument <code>ab</code>, <code>mo</code>, and <code>uti</code>: <code>as.sir(..., ab = "column1", mo = "column2", uti = "column3")</code>. This greatly improves the flexibility for users.</li>
|
||||
<li>Users can now set their own criteria (using regular expressions) as to what should be considered S, I, R, SDD, and NI.</li>
|
||||
@ -149,13 +154,13 @@
|
||||
<li>Added over 1,500 trade names for antibiotics</li>
|
||||
<li>Fix for using a manual value for <code>mo_transform</code> in <code><a href="../reference/antibiogram.html">antibiogram()</a></code>
|
||||
</li>
|
||||
<li>Fixed a bug for when <code><a href="../reference/antibiogram.html">antibiogram()</a></code> returns an empty data set</li>
|
||||
<li>Fix for mapping ‘high level’ antibiotics in <code><a href="../reference/as.ab.html">as.ab()</a></code> (amphotericin B-high, gentamicin-high, kanamycin-high, streptomycin-high, tobramycin-high)</li>
|
||||
<li>Improved overall algorithm of <code><a href="../reference/as.ab.html">as.ab()</a></code> for better performance and accuracy</li>
|
||||
<li>Improved overall algorithm of <code><a href="../reference/as.mo.html">as.mo()</a></code> for better performance and accuracy. Specifically:
|
||||
<ul><li>More weight is given to genus and species combinations in cases where the subspecies is miswritten, so that the result will be the correct genus and species</li>
|
||||
<li>Genera from the World Health Organization’s (WHO) Priority Pathogen List now have the highest prevalence</li>
|
||||
</ul></li>
|
||||
<li>Fixed a bug for when <code><a href="../reference/antibiogram.html">antibiogram()</a></code> returns an empty data set</li>
|
||||
<li>Fixed a bug for <code><a href="../reference/proportion.html">sir_confidence_interval()</a></code> when there are no isolates available</li>
|
||||
<li>Updated the prevalence calculation to include genera from the World Health Organization’s (WHO) Priority Pathogen List</li>
|
||||
<li>Improved algorithm of <code><a href="../reference/first_isolate.html">first_isolate()</a></code> when using the phenotype-based method, to prioritise records with the highest availability of SIR values</li>
|
||||
@ -167,15 +172,16 @@
|
||||
</ul></li>
|
||||
</ul></div>
|
||||
<div class="section level3">
|
||||
<h3 id="other-2-1-1-9125">Other<a class="anchor" aria-label="anchor" href="#other-2-1-1-9125"></a></h3>
|
||||
<ul><li>Greatly improved <code>vctrs</code> integration, a Tidyverse package working in the background for many Tidyverse functions. For users, this means that functions such as <code>dplyr</code>’s <code><a href="https://dplyr.tidyverse.org/reference/bind_rows.html" class="external-link">bind_rows()</a></code>, <code><a href="https://dplyr.tidyverse.org/reference/rowwise.html" class="external-link">rowwise()</a></code> and <code><a href="https://dplyr.tidyverse.org/reference/c_across.html" class="external-link">c_across()</a></code> are now supported for e.g. columns of class <code>mic</code>. Despite this, this <code>AMR</code> package is still zero-dependent on any other package, including <code>dplyr</code> and <code>vctrs</code>.</li>
|
||||
<h3 id="other-2-1-1-9133">Other<a class="anchor" aria-label="anchor" href="#other-2-1-1-9133"></a></h3>
|
||||
<ul><li>Added Dr. Larisse Bolton as contributor for her fantastic implementation of WISCA in a mathematically solid way</li>
|
||||
<li>Added Matthew Saab, Dr. Jordan Stull, and Prof. Javier Sanchez as contributors for their tremendous input on veterinary breakpoints and interpretations</li>
|
||||
<li>Greatly improved <code>vctrs</code> integration, a Tidyverse package working in the background for many Tidyverse functions. For users, this means that functions such as <code>dplyr</code>’s <code><a href="https://dplyr.tidyverse.org/reference/bind_rows.html" class="external-link">bind_rows()</a></code>, <code><a href="https://dplyr.tidyverse.org/reference/rowwise.html" class="external-link">rowwise()</a></code> and <code><a href="https://dplyr.tidyverse.org/reference/c_across.html" class="external-link">c_across()</a></code> are now supported for e.g. columns of class <code>mic</code>. Despite this, this <code>AMR</code> package is still zero-dependent on any other package, including <code>dplyr</code> and <code>vctrs</code>.</li>
|
||||
<li>Greatly updated and expanded documentation</li>
|
||||
<li>Added Larisse Bolton, Jordan Stull, Matthew Saab, and Javier Sanchez as contributors, to thank them for their valuable input</li>
|
||||
<li>Stopped support for SAS (<code>.xpt</code>) files, since their file structure and extremely inefficient and requires more disk space than GitHub allows in a single commit.</li>
|
||||
</ul></div>
|
||||
<div class="section level3">
|
||||
<h3 id="older-versions-2-1-1-9125">Older Versions<a class="anchor" aria-label="anchor" href="#older-versions-2-1-1-9125"></a></h3>
|
||||
<p>This changelog only contains changes from AMR v3.0 (October 2024) and later.</p>
|
||||
<h3 id="older-versions-2-1-1-9133">Older Versions<a class="anchor" aria-label="anchor" href="#older-versions-2-1-1-9133"></a></h3>
|
||||
<p>This changelog only contains changes from AMR v3.0 (February 2025) and later.</p>
|
||||
<ul><li>For prior v2 versions, please see <a href="https://github.com/msberends/AMR/blob/v2.1.1/NEWS.md" class="external-link">our v2 archive</a>.</li>
|
||||
<li>For prior v1 versions, please see <a href="https://github.com/msberends/AMR/blob/v1.8.2/NEWS.md" class="external-link">our v1 archive</a>.</li>
|
||||
</ul></div>
|
||||
|
||||
@ -12,7 +12,7 @@ articles:
|
||||
resistance_predict: resistance_predict.html
|
||||
welcome_to_AMR: welcome_to_AMR.html
|
||||
WHONET: WHONET.html
|
||||
last_built: 2025-01-17T11:15Z
|
||||
last_built: 2025-01-27T21:12Z
|
||||
urls:
|
||||
reference: https://msberends.github.io/AMR/reference
|
||||
article: https://msberends.github.io/AMR/articles
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -21,7 +21,7 @@ The AMR package is available in English, Chinese, Czech, Danish, Dutch, Finnish,
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -54,15 +54,9 @@
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">ab_from_text</span><span class="op">(</span></span>
|
||||
<span> <span class="va">text</span>,</span>
|
||||
<span> type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"drug"</span>, <span class="st">"dose"</span>, <span class="st">"administration"</span><span class="op">)</span>,</span>
|
||||
<span> collapse <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> translate_ab <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> thorough_search <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> info <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/interactive.html" class="external-link">interactive</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span></code></pre></div>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">ab_from_text</span><span class="op">(</span><span class="va">text</span>, type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"drug"</span>, <span class="st">"dose"</span>, <span class="st">"administration"</span><span class="op">)</span>,</span>
|
||||
<span> collapse <span class="op">=</span> <span class="cn">NULL</span>, translate_ab <span class="op">=</span> <span class="cn">FALSE</span>, thorough_search <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> info <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/interactive.html" class="external-link">interactive</a></span><span class="op">(</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -82,13 +82,8 @@
|
||||
<span></span>
|
||||
<span><span class="fu">ab_property</span><span class="op">(</span><span class="va">x</span>, property <span class="op">=</span> <span class="st">"name"</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">set_ab_names</span><span class="op">(</span></span>
|
||||
<span> <span class="va">data</span>,</span>
|
||||
<span> <span class="va">...</span>,</span>
|
||||
<span> property <span class="op">=</span> <span class="st">"name"</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> snake_case <span class="op">=</span> <span class="cn">NULL</span></span>
|
||||
<span><span class="op">)</span></span></code></pre></div>
|
||||
<span><span class="fu">set_ab_names</span><span class="op">(</span><span class="va">data</span>, <span class="va">...</span>, property <span class="op">=</span> <span class="st">"name"</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> snake_case <span class="op">=</span> <span class="cn">NULL</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -111,16 +111,16 @@
|
||||
<span class="r-in"><span></span></span>
|
||||
<span class="r-in"><span><span class="va">df</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> birth_date age age_exact age_at_y2k</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 1 1965-12-05 59 59.11781 34</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 2 1980-03-01 44 44.88219 19</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 3 1949-11-01 75 75.21096 50</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 4 1947-02-14 77 77.92329 52</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 5 1940-02-19 84 84.90959 59</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 6 1988-01-10 37 37.01918 11</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 7 1997-08-27 27 27.39178 2</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 8 1978-01-26 46 46.97534 21</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 9 1972-06-17 52 52.58630 27</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 10 1986-08-10 38 38.43836 13</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 1 1965-12-05 59 59.14521 34</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 2 1980-03-01 44 44.90959 19</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 3 1949-11-01 75 75.23836 50</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 4 1947-02-14 77 77.95068 52</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 5 1940-02-19 84 84.93699 59</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 6 1988-01-10 37 37.04658 11</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 7 1997-08-27 27 27.41918 2</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 8 1978-01-26 47 47.00274 21</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 9 1972-06-17 52 52.61370 27</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 10 1986-08-10 38 38.46575 13</span>
|
||||
</code></pre></div>
|
||||
</div>
|
||||
</main><aside class="col-md-3"><nav id="toc" aria-label="Table of contents"><h2>On this page</h2>
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
|
Before 
(image error) Size: 34 KiB After 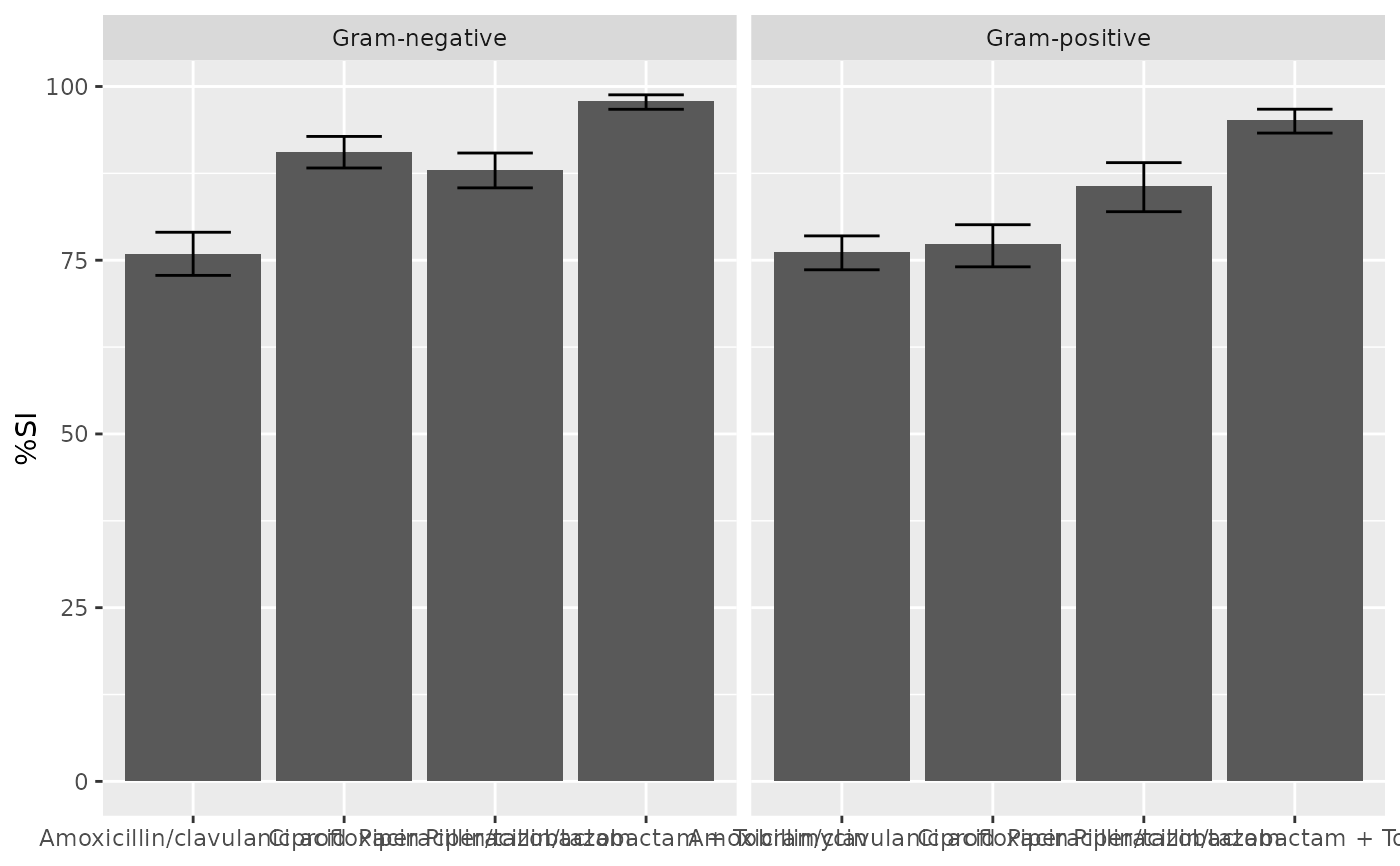
(image error) Size: 36 KiB 

|
|
Before 
(image error) Size: 29 KiB After 
(image error) Size: 31 KiB 

|
@ -1,5 +1,7 @@
|
||||
<!DOCTYPE html>
|
||||
<!-- Generated by pkgdown: do not edit by hand --><html lang="en"><head><meta http-equiv="Content-Type" content="text/html; charset=UTF-8"><meta charset="utf-8"><meta http-equiv="X-UA-Compatible" content="IE=edge"><meta name="viewport" content="width=device-width, initial-scale=1, shrink-to-fit=no"><title>Generate Traditional, Combination, Syndromic, or WISCA Antibiograms — antibiogram • AMR (for R)</title><!-- favicons --><link rel="icon" type="image/png" sizes="16x16" href="../favicon-16x16.png"><link rel="icon" type="image/png" sizes="32x32" href="../favicon-32x32.png"><link rel="apple-touch-icon" type="image/png" sizes="180x180" href="../apple-touch-icon.png"><link rel="apple-touch-icon" type="image/png" sizes="120x120" href="../apple-touch-icon-120x120.png"><link rel="apple-touch-icon" type="image/png" sizes="76x76" href="../apple-touch-icon-76x76.png"><link rel="apple-touch-icon" type="image/png" sizes="60x60" href="../apple-touch-icon-60x60.png"><script src="../deps/jquery-3.6.0/jquery-3.6.0.min.js"></script><meta name="viewport" content="width=device-width, initial-scale=1, shrink-to-fit=no"><link href="../deps/bootstrap-5.3.1/bootstrap.min.css" rel="stylesheet"><script src="../deps/bootstrap-5.3.1/bootstrap.bundle.min.js"></script><link href="../deps/Lato-0.4.9/font.css" rel="stylesheet"><link href="../deps/Fira_Code-0.4.9/font.css" rel="stylesheet"><link href="../deps/font-awesome-6.5.2/css/all.min.css" rel="stylesheet"><link href="../deps/font-awesome-6.5.2/css/v4-shims.min.css" rel="stylesheet"><script src="../deps/headroom-0.11.0/headroom.min.js"></script><script src="../deps/headroom-0.11.0/jQuery.headroom.min.js"></script><script src="../deps/bootstrap-toc-1.0.1/bootstrap-toc.min.js"></script><script src="../deps/clipboard.js-2.0.11/clipboard.min.js"></script><script src="../deps/search-1.0.0/autocomplete.jquery.min.js"></script><script src="../deps/search-1.0.0/fuse.min.js"></script><script src="../deps/search-1.0.0/mark.min.js"></script><!-- pkgdown --><script src="../pkgdown.js"></script><link href="../extra.css" rel="stylesheet"><script src="../extra.js"></script><meta property="og:title" content="Generate Traditional, Combination, Syndromic, or WISCA Antibiograms — antibiogram"><meta name="description" content="Create detailed antibiograms with options for traditional, combination, syndromic, and Bayesian WISCA methods. Based on the approaches of Klinker et al., Barbieri et al., and the Bayesian WISCA model (Weighted-Incidence Syndromic Combination Antibiogram) by Bielicki et al., this function provides flexible output formats including plots and tables, ideal for integration with R Markdown and Quarto reports."><meta property="og:description" content="Create detailed antibiograms with options for traditional, combination, syndromic, and Bayesian WISCA methods. Based on the approaches of Klinker et al., Barbieri et al., and the Bayesian WISCA model (Weighted-Incidence Syndromic Combination Antibiogram) by Bielicki et al., this function provides flexible output formats including plots and tables, ideal for integration with R Markdown and Quarto reports."><meta property="og:image" content="https://msberends.github.io/AMR/logo.svg"></head><body>
|
||||
<!-- Generated by pkgdown: do not edit by hand --><html lang="en"><head><meta http-equiv="Content-Type" content="text/html; charset=UTF-8"><meta charset="utf-8"><meta http-equiv="X-UA-Compatible" content="IE=edge"><meta name="viewport" content="width=device-width, initial-scale=1, shrink-to-fit=no"><title>Generate Traditional, Combination, Syndromic, or WISCA Antibiograms — antibiogram • AMR (for R)</title><!-- favicons --><link rel="icon" type="image/png" sizes="16x16" href="../favicon-16x16.png"><link rel="icon" type="image/png" sizes="32x32" href="../favicon-32x32.png"><link rel="apple-touch-icon" type="image/png" sizes="180x180" href="../apple-touch-icon.png"><link rel="apple-touch-icon" type="image/png" sizes="120x120" href="../apple-touch-icon-120x120.png"><link rel="apple-touch-icon" type="image/png" sizes="76x76" href="../apple-touch-icon-76x76.png"><link rel="apple-touch-icon" type="image/png" sizes="60x60" href="../apple-touch-icon-60x60.png"><script src="../deps/jquery-3.6.0/jquery-3.6.0.min.js"></script><meta name="viewport" content="width=device-width, initial-scale=1, shrink-to-fit=no"><link href="../deps/bootstrap-5.3.1/bootstrap.min.css" rel="stylesheet"><script src="../deps/bootstrap-5.3.1/bootstrap.bundle.min.js"></script><link href="../deps/Lato-0.4.9/font.css" rel="stylesheet"><link href="../deps/Fira_Code-0.4.9/font.css" rel="stylesheet"><link href="../deps/font-awesome-6.5.2/css/all.min.css" rel="stylesheet"><link href="../deps/font-awesome-6.5.2/css/v4-shims.min.css" rel="stylesheet"><script src="../deps/headroom-0.11.0/headroom.min.js"></script><script src="../deps/headroom-0.11.0/jQuery.headroom.min.js"></script><script src="../deps/bootstrap-toc-1.0.1/bootstrap-toc.min.js"></script><script src="../deps/clipboard.js-2.0.11/clipboard.min.js"></script><script src="../deps/search-1.0.0/autocomplete.jquery.min.js"></script><script src="../deps/search-1.0.0/fuse.min.js"></script><script src="../deps/search-1.0.0/mark.min.js"></script><!-- pkgdown --><script src="../pkgdown.js"></script><link href="../extra.css" rel="stylesheet"><script src="../extra.js"></script><meta property="og:title" content="Generate Traditional, Combination, Syndromic, or WISCA Antibiograms — antibiogram"><meta name="description" content="Create detailed antibiograms with options for traditional, combination, syndromic, and Bayesian WISCA methods.
|
||||
Adhering to previously described approaches (see Source) and especially the Bayesian WISCA model (Weighted-Incidence Syndromic Combination Antibiogram) by Bielicki et al., these functions provides flexible output formats including plots and tables, ideal for integration with R Markdown and Quarto reports."><meta property="og:description" content="Create detailed antibiograms with options for traditional, combination, syndromic, and Bayesian WISCA methods.
|
||||
Adhering to previously described approaches (see Source) and especially the Bayesian WISCA model (Weighted-Incidence Syndromic Combination Antibiogram) by Bielicki et al., these functions provides flexible output formats including plots and tables, ideal for integration with R Markdown and Quarto reports."><meta property="og:image" content="https://msberends.github.io/AMR/logo.svg"></head><body>
|
||||
<a href="#main" class="visually-hidden-focusable">Skip to contents</a>
|
||||
|
||||
|
||||
@ -7,7 +9,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -49,28 +51,28 @@
|
||||
</div>
|
||||
|
||||
<div class="ref-description section level2">
|
||||
<p>Create detailed antibiograms with options for traditional, combination, syndromic, and Bayesian WISCA methods. Based on the approaches of Klinker <em>et al.</em>, Barbieri <em>et al.</em>, and the Bayesian WISCA model (Weighted-Incidence Syndromic Combination Antibiogram) by Bielicki <em>et al.</em>, this function provides flexible output formats including plots and tables, ideal for integration with R Markdown and Quarto reports.</p>
|
||||
<p>Create detailed antibiograms with options for traditional, combination, syndromic, and Bayesian WISCA methods.</p>
|
||||
<p>Adhering to previously described approaches (see <em>Source</em>) and especially the Bayesian WISCA model (Weighted-Incidence Syndromic Combination Antibiogram) by Bielicki <em>et al.</em>, these functions provides flexible output formats including plots and tables, ideal for integration with R Markdown and Quarto reports.</p>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">antibiogram</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> antibiotics <span class="op">=</span> <span class="fu"><a href="https://tidyselect.r-lib.org/reference/where.html" class="external-link">where</a></span><span class="op">(</span><span class="va">is.sir</span><span class="op">)</span>,</span>
|
||||
<span> mo_transform <span class="op">=</span> <span class="st">"shortname"</span>,</span>
|
||||
<span> ab_transform <span class="op">=</span> <span class="st">"name"</span>,</span>
|
||||
<span> syndromic_group <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> add_total_n <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_all_tested <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> digits <span class="op">=</span> <span class="fl">0</span>,</span>
|
||||
<span> formatting_type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_antibiogram_formatting_type"</span>, <span class="fl">10</span><span class="op">)</span>,</span>
|
||||
<span> col_mo <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> minimum <span class="op">=</span> <span class="fl">30</span>,</span>
|
||||
<span> combine_SI <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> sep <span class="op">=</span> <span class="st">" + "</span>,</span>
|
||||
<span> info <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/interactive.html" class="external-link">interactive</a></span><span class="op">(</span><span class="op">)</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">antibiogram</span><span class="op">(</span><span class="va">x</span>, antibiotics <span class="op">=</span> <span class="fu"><a href="https://tidyselect.r-lib.org/reference/where.html" class="external-link">where</a></span><span class="op">(</span><span class="va">is.sir</span><span class="op">)</span>, mo_transform <span class="op">=</span> <span class="st">"shortname"</span>,</span>
|
||||
<span> ab_transform <span class="op">=</span> <span class="st">"name"</span>, syndromic_group <span class="op">=</span> <span class="cn">NULL</span>, add_total_n <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_all_tested <span class="op">=</span> <span class="cn">FALSE</span>, digits <span class="op">=</span> <span class="fl">0</span>,</span>
|
||||
<span> formatting_type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_antibiogram_formatting_type"</span>,</span>
|
||||
<span> <span class="fu"><a href="https://rdrr.io/r/base/ifelse.html" class="external-link">ifelse</a></span><span class="op">(</span><span class="va">wisca</span>, <span class="fl">18</span>, <span class="fl">10</span><span class="op">)</span><span class="op">)</span>, col_mo <span class="op">=</span> <span class="cn">NULL</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> minimum <span class="op">=</span> <span class="fl">30</span>, combine_SI <span class="op">=</span> <span class="cn">TRUE</span>, sep <span class="op">=</span> <span class="st">" + "</span>, wisca <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> simulations <span class="op">=</span> <span class="fl">1000</span>, conf_interval <span class="op">=</span> <span class="fl">0.95</span>, interval_side <span class="op">=</span> <span class="st">"two-tailed"</span>,</span>
|
||||
<span> info <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/interactive.html" class="external-link">interactive</a></span><span class="op">(</span><span class="op">)</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">wisca</span><span class="op">(</span><span class="va">x</span>, antibiotics <span class="op">=</span> <span class="fu"><a href="https://tidyselect.r-lib.org/reference/where.html" class="external-link">where</a></span><span class="op">(</span><span class="va">is.sir</span><span class="op">)</span>, mo_transform <span class="op">=</span> <span class="st">"shortname"</span>,</span>
|
||||
<span> ab_transform <span class="op">=</span> <span class="st">"name"</span>, syndromic_group <span class="op">=</span> <span class="cn">NULL</span>, add_total_n <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_all_tested <span class="op">=</span> <span class="cn">FALSE</span>, digits <span class="op">=</span> <span class="fl">0</span>,</span>
|
||||
<span> formatting_type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_antibiogram_formatting_type"</span>, <span class="fl">18</span><span class="op">)</span>,</span>
|
||||
<span> col_mo <span class="op">=</span> <span class="cn">NULL</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>, minimum <span class="op">=</span> <span class="fl">30</span>,</span>
|
||||
<span> combine_SI <span class="op">=</span> <span class="cn">TRUE</span>, sep <span class="op">=</span> <span class="st">" + "</span>, simulations <span class="op">=</span> <span class="fl">1000</span>,</span>
|
||||
<span> info <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/interactive.html" class="external-link">interactive</a></span><span class="op">(</span><span class="op">)</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="co"># S3 method for class 'antibiogram'</span></span>
|
||||
<span><span class="fu"><a href="plot.html">plot</a></span><span class="op">(</span><span class="va">x</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
@ -79,18 +81,15 @@
|
||||
<span><span class="fu"><a href="https://ggplot2.tidyverse.org/reference/autoplot.html" class="external-link">autoplot</a></span><span class="op">(</span><span class="va">object</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="co"># S3 method for class 'antibiogram'</span></span>
|
||||
<span><span class="fu">knit_print</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> italicise <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> na <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"knitr.kable.NA"</span>, default <span class="op">=</span> <span class="st">""</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span></code></pre></div>
|
||||
<span><span class="fu">knit_print</span><span class="op">(</span><span class="va">x</span>, italicise <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> na <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"knitr.kable.NA"</span>, default <span class="op">=</span> <span class="st">""</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="source">Source<a class="anchor" aria-label="anchor" href="#source"></a></h2>
|
||||
|
||||
<ul><li><p>Bielicki JA <em>et al.</em> (2016). <strong>Selecting appropriate empirical antibiotic regimens for paediatric bloodstream infections: application of a Bayesian decision model to local and pooled antimicrobial resistance surveillance data</strong> <em>Journal of Antimicrobial Chemotherapy</em> 71(3); <a href="https://doi.org/10.1093/jac/dkv397" class="external-link">doi:10.1093/jac/dkv397</a></p></li>
|
||||
<li><p>Bielicki JA <em>et al.</em> (2020). <strong>Evaluation of the coverage of 3 antibiotic regimens for neonatal sepsis in the hospital setting across Asian countries</strong> <em>JAMA Netw Open.</em> 3(2):e1921124; <a href="https://doi.org/10.1001.jamanetworkopen.2019.21124" class="external-link">doi:10.1001.jamanetworkopen.2019.21124</a></p></li>
|
||||
<li><p>Klinker KP <em>et al.</em> (2021). <strong>Antimicrobial stewardship and antibiograms: importance of moving beyond traditional antibiograms</strong>. <em>Therapeutic Advances in Infectious Disease</em>, May 5;8:20499361211011373; <a href="https://doi.org/10.1177/20499361211011373" class="external-link">doi:10.1177/20499361211011373</a></p></li>
|
||||
<li><p>Barbieri E <em>et al.</em> (2021). <strong>Development of a Weighted-Incidence Syndromic Combination Antibiogram (WISCA) to guide the choice of the empiric antibiotic treatment for urinary tract infection in paediatric patients: a Bayesian approach</strong> <em>Antimicrobial Resistance & Infection Control</em> May 1;10(1):74; <a href="https://doi.org/10.1186/s13756-021-00939-2" class="external-link">doi:10.1186/s13756-021-00939-2</a></p></li>
|
||||
<li><p><strong>M39 Analysis and Presentation of Cumulative Antimicrobial Susceptibility Test Data, 5th Edition</strong>, 2022, <em>Clinical and Laboratory Standards Institute (CLSI)</em>. <a href="https://clsi.org/standards/products/microbiology/documents/m39/" class="external-link">https://clsi.org/standards/products/microbiology/documents/m39/</a>.</p></li>
|
||||
@ -100,11 +99,11 @@
|
||||
|
||||
|
||||
<dl><dt id="arg-x">x<a class="anchor" aria-label="anchor" href="#arg-x"></a></dt>
|
||||
<dd><p>a <a href="https://rdrr.io/r/base/data.frame.html" class="external-link">data.frame</a> containing at least a column with microorganisms and columns with antibiotic results (class 'sir', see <code><a href="as.sir.html">as.sir()</a></code>)</p></dd>
|
||||
<dd><p>a <a href="https://rdrr.io/r/base/data.frame.html" class="external-link">data.frame</a> containing at least a column with microorganisms and columns with antimicrobial results (class 'sir', see <code><a href="as.sir.html">as.sir()</a></code>)</p></dd>
|
||||
|
||||
|
||||
<dt id="arg-antibiotics">antibiotics<a class="anchor" aria-label="anchor" href="#arg-antibiotics"></a></dt>
|
||||
<dd><p>vector of any antibiotic name or code (will be evaluated with <code><a href="as.ab.html">as.ab()</a></code>, column name of <code>x</code>, or (any combinations of) <a href="antimicrobial_class_selectors.html">antimicrobial selectors</a> such as <code><a href="antimicrobial_class_selectors.html">aminoglycosides()</a></code> or <code><a href="antimicrobial_class_selectors.html">carbapenems()</a></code>. For combination antibiograms, this can also be set to values separated with <code>"+"</code>, such as "TZP+TOB" or "cipro + genta", given that columns resembling such antibiotics exist in <code>x</code>. See <em>Examples</em>.</p></dd>
|
||||
<dd><p>vector of any antimicrobial name or code (will be evaluated with <code><a href="as.ab.html">as.ab()</a></code>, column name of <code>x</code>, or (any combinations of) <a href="antimicrobial_class_selectors.html">antimicrobial selectors</a> such as <code><a href="antimicrobial_class_selectors.html">aminoglycosides()</a></code> or <code><a href="antimicrobial_class_selectors.html">carbapenems()</a></code>. For combination antibiograms, this can also be set to values separated with <code>"+"</code>, such as "TZP+TOB" or "cipro + genta", given that columns resembling such antimicrobials exist in <code>x</code>. See <em>Examples</em>.</p></dd>
|
||||
|
||||
|
||||
<dt id="arg-mo-transform">mo_transform<a class="anchor" aria-label="anchor" href="#arg-mo-transform"></a></dt>
|
||||
@ -112,7 +111,7 @@
|
||||
|
||||
|
||||
<dt id="arg-ab-transform">ab_transform<a class="anchor" aria-label="anchor" href="#arg-ab-transform"></a></dt>
|
||||
<dd><p>a character to transform antibiotic input - must be one of the column names of the <a href="antibiotics.html">antibiotics</a> data set (defaults to <code>"name"</code>): "ab", "cid", "name", "group", "atc", "atc_group1", "atc_group2", "abbreviations", "synonyms", "oral_ddd", "oral_units", "iv_ddd", "iv_units", or "loinc". Can also be <code>NULL</code> to not transform the input.</p></dd>
|
||||
<dd><p>a character to transform antimicrobial input - must be one of the column names of the <a href="antibiotics.html">antibiotics</a> data set (defaults to <code>"name"</code>): "ab", "cid", "name", "group", "atc", "atc_group1", "atc_group2", "abbreviations", "synonyms", "oral_ddd", "oral_units", "iv_ddd", "iv_units", or "loinc". Can also be <code>NULL</code> to not transform the input.</p></dd>
|
||||
|
||||
|
||||
<dt id="arg-syndromic-group">syndromic_group<a class="anchor" aria-label="anchor" href="#arg-syndromic-group"></a></dt>
|
||||
@ -120,11 +119,11 @@
|
||||
|
||||
|
||||
<dt id="arg-add-total-n">add_total_n<a class="anchor" aria-label="anchor" href="#arg-add-total-n"></a></dt>
|
||||
<dd><p>a <a href="https://rdrr.io/r/base/logical.html" class="external-link">logical</a> to indicate whether total available numbers per pathogen should be added to the table (default is <code>TRUE</code>). This will add the lowest and highest number of available isolate per antibiotic (e.g, if for <em>E. coli</em> 200 isolates are available for ciprofloxacin and 150 for amoxicillin, the returned number will be "150-200").</p></dd>
|
||||
<dd><p>a <a href="https://rdrr.io/r/base/logical.html" class="external-link">logical</a> to indicate whether total available numbers per pathogen should be added to the table (default is <code>TRUE</code>). This will add the lowest and highest number of available isolates per antimicrobial (e.g, if for <em>E. coli</em> 200 isolates are available for ciprofloxacin and 150 for amoxicillin, the returned number will be "150-200").</p></dd>
|
||||
|
||||
|
||||
<dt id="arg-only-all-tested">only_all_tested<a class="anchor" aria-label="anchor" href="#arg-only-all-tested"></a></dt>
|
||||
<dd><p>(for combination antibiograms): a <a href="https://rdrr.io/r/base/logical.html" class="external-link">logical</a> to indicate that isolates must be tested for all antibiotics, see <em>Details</em></p></dd>
|
||||
<dd><p>(for combination antibiograms): a <a href="https://rdrr.io/r/base/logical.html" class="external-link">logical</a> to indicate that isolates must be tested for all antimicrobials, see <em>Details</em></p></dd>
|
||||
|
||||
|
||||
<dt id="arg-digits">digits<a class="anchor" aria-label="anchor" href="#arg-digits"></a></dt>
|
||||
@ -132,7 +131,7 @@
|
||||
|
||||
|
||||
<dt id="arg-formatting-type">formatting_type<a class="anchor" aria-label="anchor" href="#arg-formatting-type"></a></dt>
|
||||
<dd><p>numeric value (1–12) indicating how the 'cells' of the antibiogram table should be formatted. See <em>Details</em> > <em>Formatting Type</em> for a list of options.</p></dd>
|
||||
<dd><p>numeric value (1–22 for WISCA, 1-12 for non-WISCA) indicating how the 'cells' of the antibiogram table should be formatted. See <em>Details</em> > <em>Formatting Type</em> for a list of options.</p></dd>
|
||||
|
||||
|
||||
<dt id="arg-col-mo">col_mo<a class="anchor" aria-label="anchor" href="#arg-col-mo"></a></dt>
|
||||
@ -152,7 +151,23 @@
|
||||
|
||||
|
||||
<dt id="arg-sep">sep<a class="anchor" aria-label="anchor" href="#arg-sep"></a></dt>
|
||||
<dd><p>a separating character for antibiotic columns in combination antibiograms</p></dd>
|
||||
<dd><p>a separating character for antimicrobial columns in combination antibiograms</p></dd>
|
||||
|
||||
|
||||
<dt id="arg-wisca">wisca<a class="anchor" aria-label="anchor" href="#arg-wisca"></a></dt>
|
||||
<dd><p>a <a href="https://rdrr.io/r/base/logical.html" class="external-link">logical</a> to indicate whether a Weighted-Incidence Syndromic Combination Antibiogram (WISCA) must be generated (default is <code>FALSE</code>). This will use a Bayesian hierarchical model to estimate regimen coverage probabilities using Montecarlo simulations. Set <code>simulations</code> to adjust.</p></dd>
|
||||
|
||||
|
||||
<dt id="arg-simulations">simulations<a class="anchor" aria-label="anchor" href="#arg-simulations"></a></dt>
|
||||
<dd><p>(for WISCA) a numerical value to set the number of Montecarlo simulations</p></dd>
|
||||
|
||||
|
||||
<dt id="arg-conf-interval">conf_interval<a class="anchor" aria-label="anchor" href="#arg-conf-interval"></a></dt>
|
||||
<dd><p>(for WISCA) a numerical value to set confidence interval (default is <code>0.95</code>)</p></dd>
|
||||
|
||||
|
||||
<dt id="arg-interval-side">interval_side<a class="anchor" aria-label="anchor" href="#arg-interval-side"></a></dt>
|
||||
<dd><p>(for WISCA) the side of the confidence interval, either <code>"two-tailed"</code> (default), <code>"left"</code> or <code>"right"</code></p></dd>
|
||||
|
||||
|
||||
<dt id="arg-info">info<a class="anchor" aria-label="anchor" href="#arg-info"></a></dt>
|
||||
@ -178,11 +193,13 @@
|
||||
<div class="section level2">
|
||||
<h2 id="details">Details<a class="anchor" aria-label="anchor" href="#details"></a></h2>
|
||||
<p>This function returns a table with values between 0 and 100 for <em>susceptibility</em>, not resistance.</p>
|
||||
<p><strong>Remember that you should filter your data to let it contain only first isolates!</strong> This is needed to exclude duplicates and to reduce selection bias. Use <code><a href="first_isolate.html">first_isolate()</a></code> to determine them in your data set with one of the four available algorithms.</p><div class="section">
|
||||
<p><strong>Remember that you should filter your data to let it contain only first isolates!</strong> This is needed to exclude duplicates and to reduce selection bias. Use <code><a href="first_isolate.html">first_isolate()</a></code> to determine them in your data set with one of the four available algorithms.</p>
|
||||
<p>For estimating antimicrobial coverage, especially when creating a WISCA, the outcome might become more reliable by only including the top <em>n</em> species encountered in the data. You can filter on this top <em>n</em> using <code><a href="top_n_microorganisms.html">top_n_microorganisms()</a></code>. For example, use <code>top_n_microorganisms(your_data, n = 10)</code> as a pre-processing step to only include the top 10 species in the data.</p>
|
||||
<p>The numeric values of an antibiogram are stored in a long format as the <a href="https://rdrr.io/r/base/attributes.html" class="external-link">attribute</a> <code>long_numeric</code>. You can retrieve them using <code>attributes(x)$long_numeric</code>, where <code>x</code> is the outcome of <code>antibiogram()</code> or <code>wisca()</code>. This is ideal for e.g. advanced plotting.</p><div class="section">
|
||||
<h3 id="formatting-type">Formatting Type<a class="anchor" aria-label="anchor" href="#formatting-type"></a></h3>
|
||||
|
||||
|
||||
<p>The formatting of the 'cells' of the table can be set with the argument <code>formatting_type</code>. In these examples, <code>5</code> is the susceptibility percentage, <code>15</code> the numerator, and <code>300</code> the denominator:</p><ol><li><p>5</p></li>
|
||||
<p>The formatting of the 'cells' of the table can be set with the argument <code>formatting_type</code>. In these examples, <code>5</code> is the susceptibility percentage (for WISCA: <code>4-6</code> indicates the confidence level), <code>15</code> the numerator, and <code>300</code> the denominator:</p><ol><li><p>5</p></li>
|
||||
<li><p>15</p></li>
|
||||
<li><p>300</p></li>
|
||||
<li><p>15/300</p></li>
|
||||
@ -191,20 +208,32 @@
|
||||
<li><p>5 (N=300)</p></li>
|
||||
<li><p>5% (N=300)</p></li>
|
||||
<li><p>5 (15/300)</p></li>
|
||||
<li><p>5% (15/300)</p></li>
|
||||
<li><p>5% (15/300) - <strong>default for non-WISCA</strong></p></li>
|
||||
<li><p>5 (N=15/300)</p></li>
|
||||
<li><p>5% (N=15/300)</p></li>
|
||||
</ol><p>The default is <code>10</code>, which can be set globally with the package option <code><a href="AMR-options.html">AMR_antibiogram_formatting_type</a></code>, e.g. <code>options(AMR_antibiogram_formatting_type = 5)</code>.</p>
|
||||
<p>Set <code>digits</code> (defaults to <code>0</code>) to alter the rounding of the susceptibility percentage.</p>
|
||||
<li><p>5% (N=15/300)</p>
|
||||
<p>Additional options for WISCA (using <code>antibiogram(..., wisca = TRUE)</code> or <code>wisca()</code>):</p></li>
|
||||
<li><p>5 (4-6)</p></li>
|
||||
<li><p>5% (4-6%)</p></li>
|
||||
<li><p>5 (4-6,300)</p></li>
|
||||
<li><p>5% (4-6%,300)</p></li>
|
||||
<li><p>5 (4-6,N=300)</p></li>
|
||||
<li><p>5% (4-6%,N=300) - <strong>default for WISCA</strong></p></li>
|
||||
<li><p>5 (4-6,15/300)</p></li>
|
||||
<li><p>5% (4-6%,15/300)</p></li>
|
||||
<li><p>5 (4-6,N=15/300)</p></li>
|
||||
<li><p>5% (4-6%,N=15/300)</p></li>
|
||||
</ol><p>The default is <code>18</code> for WISCA and <code>10</code> for non-WISCA, which can be set globally with the package option <code><a href="AMR-options.html">AMR_antibiogram_formatting_type</a></code>, e.g. <code>options(AMR_antibiogram_formatting_type = 5)</code>.</p>
|
||||
<p>Set <code>digits</code> (defaults to <code>0</code>) to alter the rounding of the susceptibility percentages.</p>
|
||||
</div>
|
||||
|
||||
<div class="section">
|
||||
<h3 id="antibiogram-types">Antibiogram Types<a class="anchor" aria-label="anchor" href="#antibiogram-types"></a></h3>
|
||||
|
||||
|
||||
<p>There are four antibiogram types, as summarised by Klinker <em>et al.</em> (2021, <a href="https://doi.org/10.1177/20499361211011373" class="external-link">doi:10.1177/20499361211011373</a>
|
||||
), and they are all supported by <code>antibiogram()</code>. Use WISCA whenever possible, since it provides precise coverage estimates by accounting for pathogen incidence and antimicrobial susceptibility. See the section <em>Why Use WISCA?</em> on this page.</p>
|
||||
<p>The four antibiogram types:</p><ol><li><p><strong>Traditional Antibiogram</strong></p>
|
||||
<p>There are various antibiogram types, as summarised by Klinker <em>et al.</em> (2021, <a href="https://doi.org/10.1177/20499361211011373" class="external-link">doi:10.1177/20499361211011373</a>
|
||||
), and they are all supported by <code>antibiogram()</code>.</p>
|
||||
<p><strong>Use WISCA whenever possible</strong>, since it provides more precise coverage estimates by accounting for pathogen incidence and antimicrobial susceptibility, as has been shown by Bielicki <em>et al.</em> (2020, <a href="https://doi.org/10.1001.jamanetworkopen.2019.21124" class="external-link">doi:10.1001.jamanetworkopen.2019.21124</a>
|
||||
). See the section <em>Why Use WISCA?</em> on this page.</p><ol><li><p><strong>Traditional Antibiogram</strong></p>
|
||||
<p>Case example: Susceptibility of <em>Pseudomonas aeruginosa</em> to piperacillin/tazobactam (TZP)</p>
|
||||
<p>Code example:</p>
|
||||
<p></p><div class="sourceCode r"><pre><code><span><span class="fu"><a href="../reference/antibiogram.html">antibiogram</a></span><span class="op">(</span><span class="va">your_data</span>,</span>
|
||||
@ -221,25 +250,28 @@
|
||||
<span> antibiotics <span class="op">=</span> <span class="fu"><a href="../reference/antimicrobial_class_selectors.html">penicillins</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> syndromic_group <span class="op">=</span> <span class="st">"ward"</span><span class="op">)</span></span></code></pre><p></p></div></li>
|
||||
<li><p><strong>Weighted-Incidence Syndromic Combination Antibiogram (WISCA)</strong></p>
|
||||
<p>WISCA enhances empirical antibiotic selection by weighting the incidence of pathogens in specific clinical syndromes and combining them with their susceptibility data. It provides an estimation of regimen coverage by aggregating pathogen incidences and susceptibilities across potential causative organisms. See also the section <em>Why Use WISCA?</em> on this page.</p>
|
||||
<p>Case example: Susceptibility of <em>Pseudomonas aeruginosa</em> to TZP among respiratory specimens (obtained among ICU patients only) for male patients age >=65 years with heart failure</p>
|
||||
<p>WISCA can be applied to any antibiogram, see the section <em>Why Use WISCA?</em> on this page for more information.</p>
|
||||
<p>Code example:</p>
|
||||
<p></p><div class="sourceCode r"><pre><code><span><span class="kw"><a href="https://rdrr.io/r/base/library.html" class="external-link">library</a></span><span class="op">(</span><span class="va"><a href="https://dplyr.tidyverse.org" class="external-link">dplyr</a></span><span class="op">)</span></span>
|
||||
<span><span class="va">your_data</span> <span class="op"><a href="https://magrittr.tidyverse.org/reference/pipe.html" class="external-link">%>%</a></span></span>
|
||||
<span> <span class="fu"><a href="https://dplyr.tidyverse.org/reference/filter.html" class="external-link">filter</a></span><span class="op">(</span><span class="va">ward</span> <span class="op">==</span> <span class="st">"ICU"</span> <span class="op">&</span> <span class="va">specimen_type</span> <span class="op">==</span> <span class="st">"Respiratory"</span><span class="op">)</span> <span class="op"><a href="https://magrittr.tidyverse.org/reference/pipe.html" class="external-link">%>%</a></span></span>
|
||||
<span> <span class="fu"><a href="../reference/antibiogram.html">antibiogram</a></span><span class="op">(</span>antibiotics <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"TZP"</span>, <span class="st">"TZP+TOB"</span>, <span class="st">"TZP+GEN"</span><span class="op">)</span>,</span>
|
||||
<span> syndromic_group <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/ifelse.html" class="external-link">ifelse</a></span><span class="op">(</span><span class="va">.</span><span class="op">$</span><span class="va">age</span> <span class="op">>=</span> <span class="fl">65</span> <span class="op">&</span></span>
|
||||
<span> <span class="va">.</span><span class="op">$</span><span class="va">gender</span> <span class="op">==</span> <span class="st">"Male"</span> <span class="op">&</span></span>
|
||||
<span> <span class="va">.</span><span class="op">$</span><span class="va">condition</span> <span class="op">==</span> <span class="st">"Heart Disease"</span>,</span>
|
||||
<span> <span class="st">"Study Group"</span>, <span class="st">"Control Group"</span><span class="op">)</span><span class="op">)</span></span></code></pre><p></p></div>
|
||||
<p></p><div class="sourceCode r"><pre><code><span><span class="fu"><a href="../reference/antibiogram.html">antibiogram</a></span><span class="op">(</span><span class="va">your_data</span>,</span>
|
||||
<span> antibiotics <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"TZP"</span>, <span class="st">"TZP+TOB"</span>, <span class="st">"TZP+GEN"</span><span class="op">)</span>,</span>
|
||||
<span> wisca <span class="op">=</span> <span class="cn">TRUE</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="co"># this is equal to:</span></span>
|
||||
<span><span class="fu"><a href="../reference/antibiogram.html">wisca</a></span><span class="op">(</span><span class="va">your_data</span>,</span>
|
||||
<span> antibiotics <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"TZP"</span>, <span class="st">"TZP+TOB"</span>, <span class="st">"TZP+GEN"</span><span class="op">)</span><span class="op">)</span></span></code></pre><p></p></div>
|
||||
<p>WISCA uses a sophisticated Bayesian decision model to combine both local and pooled antimicrobial resistance data. This approach not only evaluates local patterns but can also draw on multi-centre datasets to improve regimen accuracy, even in low-incidence infections like paediatric bloodstream infections (BSIs).</p></li>
|
||||
</ol></div>
|
||||
</ol><p>Grouped <a href="https://tibble.tidyverse.org/reference/tibble.html" class="external-link">tibbles</a> can also be used to calculate susceptibilities over various groups.</p>
|
||||
<p>Code example:</p>
|
||||
<p></p><div class="sourceCode r"><pre><code><span><span class="va">your_data</span> <span class="op"><a href="https://magrittr.tidyverse.org/reference/pipe.html" class="external-link">%>%</a></span></span>
|
||||
<span> <span class="fu"><a href="https://dplyr.tidyverse.org/reference/group_by.html" class="external-link">group_by</a></span><span class="op">(</span><span class="va">has_sepsis</span>, <span class="va">is_neonate</span>, <span class="va">sex</span><span class="op">)</span> <span class="op"><a href="https://magrittr.tidyverse.org/reference/pipe.html" class="external-link">%>%</a></span></span>
|
||||
<span> <span class="fu"><a href="../reference/antibiogram.html">wisca</a></span><span class="op">(</span>antibiotics <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"TZP"</span>, <span class="st">"TZP+TOB"</span>, <span class="st">"TZP+GEN"</span><span class="op">)</span><span class="op">)</span></span></code></pre><p></p></div>
|
||||
</div>
|
||||
|
||||
<div class="section">
|
||||
<h3 id="inclusion-in-combination-antibiogram-and-syndromic-antibiogram">Inclusion in Combination Antibiogram and Syndromic Antibiogram<a class="anchor" aria-label="anchor" href="#inclusion-in-combination-antibiogram-and-syndromic-antibiogram"></a></h3>
|
||||
|
||||
|
||||
<p>Note that for types 2 and 3 (Combination Antibiogram and Syndromic Antibiogram), it is important to realise that susceptibility can be calculated in two ways, which can be set with the <code>only_all_tested</code> argument (default is <code>FALSE</code>). See this example for two antibiotics, Drug A and Drug B, about how <code>antibiogram()</code> works to calculate the %SI:</p>
|
||||
<p>Note that for types 2 and 3 (Combination Antibiogram and Syndromic Antibiogram), it is important to realise that susceptibility can be calculated in two ways, which can be set with the <code>only_all_tested</code> argument (default is <code>FALSE</code>). See this example for two antimicrobials, Drug A and Drug B, about how <code>antibiogram()</code> works to calculate the %SI:</p>
|
||||
<p></p><div class="sourceCode"><pre><code><span id="cb1-1"><a href="#cb1-1" tabindex="-1"></a><span class="sc">--------------------------------------------------------------------</span></span>
|
||||
<span id="cb1-2"><a href="#cb1-2" tabindex="-1"></a> only_all_tested <span class="ot">=</span> <span class="cn">FALSE</span> only_all_tested <span class="ot">=</span> <span class="cn">TRUE</span></span>
|
||||
<span id="cb1-3"><a href="#cb1-3" tabindex="-1"></a> <span class="sc">-----------------------</span> <span class="sc">-----------------------</span></span>
|
||||
@ -272,12 +304,39 @@
|
||||
<h2 id="why-use-wisca-">Why Use WISCA?<a class="anchor" aria-label="anchor" href="#why-use-wisca-"></a></h2>
|
||||
|
||||
|
||||
<p>WISCA is a powerful tool for guiding empirical antibiotic therapy because it provides precise coverage estimates by accounting for pathogen incidence and antimicrobial susceptibility. This is particularly important in empirical treatment, where the causative pathogen is often unknown at the outset. Traditional antibiograms do not reflect the weighted likelihood of specific pathogens based on clinical syndromes, which can lead to suboptimal treatment choices.</p>
|
||||
<p>The Bayesian WISCA, as described by Bielicki <em>et al.</em> (2016), improves on earlier methods by handling uncertainties common in smaller datasets, such as low-incidence infections. This method offers a significant advantage by:</p><ol><li><p>Pooling Data from Multiple Sources:<br> WISCA uses pooled data from multiple hospitals or surveillance sources to overcome limitations of small sample sizes at individual institutions, allowing for more confident selection of narrow-spectrum antibiotics or combinations.</p></li>
|
||||
<li><p>Bayesian Framework:<br> The Bayesian decision tree model accounts for both local data and prior knowledge (such as inherent resistance patterns) to estimate regimen coverage. It allows for a more precise estimation of coverage, even in cases where susceptibility data is missing or incomplete.</p></li>
|
||||
<li><p>Incorporating Pathogen and Regimen Uncertainty:<br> WISCA allows clinicians to see the likelihood that an empirical regimen will be effective against all relevant pathogens, taking into account uncertainties related to both pathogen prevalence and antimicrobial resistance. This leads to better-informed, data-driven clinical decisions.</p></li>
|
||||
<li><p>Scenarios for Optimising Treatment:<br> For hospitals or settings with low-incidence infections, WISCA helps determine whether local data is sufficient or if pooling with external data is necessary. It also identifies statistically significant differences or similarities between antibiotic regimens, enabling clinicians to choose optimal therapies with greater confidence.</p></li>
|
||||
</ol><p>WISCA is essential in optimising empirical treatment by shifting away from broad-spectrum antibiotics, which are often overused in empirical settings. By offering precise estimates based on syndromic patterns and pooled data, WISCA supports antimicrobial stewardship by guiding more targeted therapy, reducing unnecessary broad-spectrum use, and combating the rise of antimicrobial resistance.</p>
|
||||
<p>WISCA, as outlined by Barbieri <em>et al.</em> (<a href="https://doi.org/10.1186/s13756-021-00939-2" class="external-link">doi:10.1186/s13756-021-00939-2</a>
|
||||
), stands for
|
||||
Weighted-Incidence Syndromic Combination Antibiogram, which estimates the probability
|
||||
of adequate empirical antimicrobial regimen coverage for specific infection syndromes.
|
||||
This method leverages a Bayesian hierarchical logistic regression framework with random
|
||||
effects for pathogens and regimens, enabling robust estimates in the presence of sparse
|
||||
data.</p>
|
||||
<p>The Bayesian model assumes conjugate priors for parameter estimation. For example, the
|
||||
coverage probability $theta$ for a given antimicrobial regimen
|
||||
is modeled using a Beta distribution as a prior:</p>
|
||||
<p><img src="figures/beta_prior.png" width="300" alt="Beta prior"></p>
|
||||
<p>where \($alpha$_0\) and \($beta$_0\) represent prior successes and failures, respectively,
|
||||
informed by expert knowledge or weakly informative priors (e.g., \($alpha$_0 = 1, $beta$_0 = 1\)).</p>
|
||||
<p>The likelihood function is constructed based on observed data, where the number of covered
|
||||
cases for a regimen follows a binomial distribution:</p>
|
||||
<p><img src="figures/binomial_likelihood.png" width="300" alt="Binomial likelihood"></p>
|
||||
<p>Posterior parameter estimates are obtained by combining the prior and likelihood using
|
||||
Bayes' theorem. The posterior distribution of \($theta$\) is also a Beta distribution:</p>
|
||||
<p><img src="figures/posterior_beta.png" width="300" alt="Beta posterior"></p>
|
||||
<p>For hierarchical modeling, pathogen-level effects (e.g., differences in resistance
|
||||
patterns) and regimen-level effects are modelled using Gaussian priors on log-odds.
|
||||
This hierarchical structure ensures partial pooling of estimates across groups,
|
||||
improving stability in strata with small sample sizes. The model is implemented using
|
||||
Hamiltonian Monte Carlo (HMC) sampling.</p>
|
||||
<p>Stratified results are provided based on covariates such as age, sex, and clinical
|
||||
complexity (e.g., prior antimicrobial treatments or renal/urological comorbidities).
|
||||
For example, posterior odds ratios (ORs) are derived to quantify the effect of these
|
||||
covariates on coverage probabilities:</p>
|
||||
<p><img src="figures/odds_ratio.png" width="300" alt="Odds ratio formula"></p>
|
||||
<p>By combining empirical data with prior knowledge, WISCA overcomes the limitations
|
||||
of traditional combination antibiograms, offering disease-specific, patient-stratified
|
||||
estimates with robust uncertainty quantification. This tool is invaluable for antimicrobial
|
||||
stewardship programs and empirical treatment guideline refinement.</p>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
@ -312,36 +371,10 @@
|
||||
<span class="r-in"><span><span class="fu">antibiogram</span><span class="op">(</span><span class="va">example_isolates</span>,</span></span>
|
||||
<span class="r-in"><span> antibiotics <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="fu"><a href="antimicrobial_class_selectors.html">aminoglycosides</a></span><span class="op">(</span><span class="op">)</span>, <span class="fu"><a href="antimicrobial_class_selectors.html">carbapenems</a></span><span class="op">(</span><span class="op">)</span><span class="op">)</span></span></span>
|
||||
<span class="r-in"><span><span class="op">)</span></span></span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> ℹ The function aminoglycosides() should be used inside a dplyr verb or</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> data.frame call, e.g.:</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% select(aminoglycosides())</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% select(column_a, column_b, aminoglycosides())</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% filter(any(aminoglycosides() == "R"))</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data[, aminoglycosides()]</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data[, c("column_a", "column_b", aminoglycosides())]</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> </span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> Now returning a vector of all possible antimicrobials that</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> aminoglycosides() can select.</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> ℹ The function carbapenems() should be used inside a dplyr verb or</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> data.frame call, e.g.:</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% select(carbapenems())</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% select(column_a, column_b, carbapenems())</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% filter(any(carbapenems() == "R"))</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data[, carbapenems()]</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data[, c("column_a", "column_b", carbapenems())]</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> </span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> Now returning a vector of all possible antimicrobials that carbapenems()</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> can select.</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> <span class="warning">Warning: </span>The following antibiotics were not available and ignored:</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> amikacin/fosfomycin, apramycin, arbekacin, astromicin, bekanamycin,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> biapenem, dibekacin, doripenem, ertapenem, framycetin, gentamicin-high,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> habekacin, hygromycin, imipenem/EDTA, imipenem/relebactam, isepamicin,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> kanamycin-high, kanamycin/cephalexin, meropenem/nacubactam,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> meropenem/vaborbactam, micronomicin, neomycin, netilmicin, panipenem,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> pentisomicin, plazomicin, propikacin, razupenem, ribostamycin, ritipenem,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> ritipenem acoxil, sisomicin, streptoduocin, streptomycin,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> streptomycin-high, tebipenem, and tobramycin-high</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># An Antibiogram: 10 × 7</span></span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> ℹ For aminoglycosides() using columns 'GEN' (gentamicin), 'TOB'</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> (tobramycin), 'AMK' (amikacin), and 'KAN' (kanamycin)</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> ℹ For carbapenems() using columns 'IPM' (imipenem) and 'MEM' (meropenem)</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># An Antibiogram (non-WISCA): 10 × 7</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> Pathogen Amikacin Gentamicin Imipenem Kanamycin Meropenem Tobramycin</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">*</span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 1</span> CoNS 0% (0/43) 86% (267/… 52% (25… 0% (0/43) 52% (25/… 22% (12/5…</span>
|
||||
@ -362,23 +395,9 @@
|
||||
<span class="r-in"><span> ab_transform <span class="op">=</span> <span class="st">"atc"</span>,</span></span>
|
||||
<span class="r-in"><span> mo_transform <span class="op">=</span> <span class="st">"gramstain"</span></span></span>
|
||||
<span class="r-in"><span><span class="op">)</span></span></span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> ℹ The function aminoglycosides() should be used inside a dplyr verb or</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> data.frame call, e.g.:</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% select(aminoglycosides())</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% select(column_a, column_b, aminoglycosides())</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% filter(any(aminoglycosides() == "R"))</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data[, aminoglycosides()]</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data[, c("column_a", "column_b", aminoglycosides())]</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> </span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> Now returning a vector of all possible antimicrobials that</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> aminoglycosides() can select.</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> <span class="warning">Warning: </span>The following antibiotics were not available and ignored:</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> amikacin/fosfomycin, apramycin, arbekacin, astromicin, bekanamycin,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> dibekacin, framycetin, gentamicin-high, habekacin, hygromycin, isepamicin,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> kanamycin-high, kanamycin/cephalexin, micronomicin, neomycin, netilmicin,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> pentisomicin, plazomicin, propikacin, ribostamycin, sisomicin,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> streptoduocin, streptomycin, streptomycin-high, and tobramycin-high</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># An Antibiogram: 2 × 5</span></span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> ℹ For aminoglycosides() using columns 'GEN' (gentamicin), 'TOB'</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> (tobramycin), 'AMK' (amikacin), and 'KAN' (kanamycin)</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># An Antibiogram (non-WISCA): 2 × 5</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> Pathogen J01GB01 J01GB03 J01GB04 J01GB06 </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">*</span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">1</span> Gram-negative 96% (658/686) 96% (659/684) 0% (0/35) 98% (251/256)</span>
|
||||
@ -391,21 +410,8 @@
|
||||
<span class="r-in"><span> ab_transform <span class="op">=</span> <span class="st">"name"</span>,</span></span>
|
||||
<span class="r-in"><span> mo_transform <span class="op">=</span> <span class="st">"name"</span></span></span>
|
||||
<span class="r-in"><span><span class="op">)</span></span></span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> ℹ The function carbapenems() should be used inside a dplyr verb or</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> data.frame call, e.g.:</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% select(carbapenems())</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% select(column_a, column_b, carbapenems())</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% filter(any(carbapenems() == "R"))</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data[, carbapenems()]</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data[, c("column_a", "column_b", carbapenems())]</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> </span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> Now returning a vector of all possible antimicrobials that carbapenems()</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> can select.</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> <span class="warning">Warning: </span>The following antibiotics were not available and ignored: biapenem,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> doripenem, ertapenem, imipenem/EDTA, imipenem/relebactam,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> meropenem/nacubactam, meropenem/vaborbactam, panipenem, razupenem,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> ritipenem, ritipenem acoxil, and tebipenem</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># An Antibiogram: 5 × 3</span></span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> ℹ For carbapenems() using columns 'IPM' (imipenem) and 'MEM' (meropenem)</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># An Antibiogram (non-WISCA): 5 × 3</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> Pathogen Imipenem Meropenem </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">*</span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">1</span> Coagulase-negative Staphylococcus (CoNS) 52% (25/48) 52% (25/48) </span>
|
||||
@ -424,7 +430,7 @@
|
||||
<span class="r-in"><span> antibiotics <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"TZP"</span>, <span class="st">"TZP+TOB"</span>, <span class="st">"TZP+GEN"</span><span class="op">)</span>,</span></span>
|
||||
<span class="r-in"><span> mo_transform <span class="op">=</span> <span class="st">"gramstain"</span></span></span>
|
||||
<span class="r-in"><span><span class="op">)</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># An Antibiogram: 2 × 4</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># An Antibiogram (non-WISCA): 2 × 4</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> Pathogen Piperacillin/tazobac…¹ Piperacillin/tazobac…² Piperacillin/tazobac…³</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">*</span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">1</span> Gram-neg… 88% (565/641) 99% (681/691) 98% (679/693) </span>
|
||||
@ -442,7 +448,7 @@
|
||||
<span class="r-in"><span> ab_transform <span class="op">=</span> <span class="st">"name"</span>,</span></span>
|
||||
<span class="r-in"><span> sep <span class="op">=</span> <span class="st">" & "</span></span></span>
|
||||
<span class="r-in"><span><span class="op">)</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># An Antibiogram: 2 × 3</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># An Antibiogram (non-WISCA): 2 × 3</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> Pathogen Ciprofloxacin `Ciprofloxacin & Gentamicin`</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">*</span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">1</span> Gram-negative 91% (621/684) 99% (684/694) </span>
|
||||
@ -458,36 +464,10 @@
|
||||
<span class="r-in"><span> antibiotics <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="fu"><a href="antimicrobial_class_selectors.html">aminoglycosides</a></span><span class="op">(</span><span class="op">)</span>, <span class="fu"><a href="antimicrobial_class_selectors.html">carbapenems</a></span><span class="op">(</span><span class="op">)</span><span class="op">)</span>,</span></span>
|
||||
<span class="r-in"><span> syndromic_group <span class="op">=</span> <span class="st">"ward"</span></span></span>
|
||||
<span class="r-in"><span><span class="op">)</span></span></span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> ℹ The function aminoglycosides() should be used inside a dplyr verb or</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> data.frame call, e.g.:</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% select(aminoglycosides())</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% select(column_a, column_b, aminoglycosides())</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% filter(any(aminoglycosides() == "R"))</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data[, aminoglycosides()]</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data[, c("column_a", "column_b", aminoglycosides())]</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> </span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> Now returning a vector of all possible antimicrobials that</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> aminoglycosides() can select.</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> ℹ The function carbapenems() should be used inside a dplyr verb or</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> data.frame call, e.g.:</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% select(carbapenems())</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% select(column_a, column_b, carbapenems())</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% filter(any(carbapenems() == "R"))</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data[, carbapenems()]</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data[, c("column_a", "column_b", carbapenems())]</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> </span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> Now returning a vector of all possible antimicrobials that carbapenems()</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> can select.</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> <span class="warning">Warning: </span>The following antibiotics were not available and ignored:</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> amikacin/fosfomycin, apramycin, arbekacin, astromicin, bekanamycin,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> biapenem, dibekacin, doripenem, ertapenem, framycetin, gentamicin-high,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> habekacin, hygromycin, imipenem/EDTA, imipenem/relebactam, isepamicin,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> kanamycin-high, kanamycin/cephalexin, meropenem/nacubactam,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> meropenem/vaborbactam, micronomicin, neomycin, netilmicin, panipenem,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> pentisomicin, plazomicin, propikacin, razupenem, ribostamycin, ritipenem,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> ritipenem acoxil, sisomicin, streptoduocin, streptomycin,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> streptomycin-high, tebipenem, and tobramycin-high</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># An Antibiogram: 14 × 8</span></span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> ℹ For aminoglycosides() using columns 'GEN' (gentamicin), 'TOB'</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> (tobramycin), 'AMK' (amikacin), and 'KAN' (kanamycin)</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> ℹ For carbapenems() using columns 'IPM' (imipenem) and 'MEM' (meropenem)</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># An Antibiogram (non-WISCA): 14 × 8</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> `Syndromic Group` Pathogen Amikacin Gentamicin Imipenem Kanamycin Meropenem</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">*</span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 1</span> Clinical CoNS <span style="color: #BB0000;">NA</span> 89% (183/… 57% (20… <span style="color: #BB0000;">NA</span> 57% (20/…</span>
|
||||
@ -522,23 +502,9 @@
|
||||
<span class="r-in"><span> <span class="op">)</span>,</span></span>
|
||||
<span class="r-in"><span> language <span class="op">=</span> <span class="st">"es"</span></span></span>
|
||||
<span class="r-in"><span><span class="op">)</span></span></span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> ℹ The function aminoglycosides() should be used inside a dplyr verb or</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> data.frame call, e.g.:</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% select(aminoglycosides())</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% select(column_a, column_b, aminoglycosides())</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% filter(any(aminoglycosides() == "R"))</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data[, aminoglycosides()]</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data[, c("column_a", "column_b", aminoglycosides())]</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> </span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> Now returning a vector of all possible antimicrobials that</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> aminoglycosides() can select.</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> <span class="warning">Warning: </span>The following antibiotics were not available and ignored:</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> amikacin/fosfomycin, apramycin, arbekacin, astromicin, bekanamycin,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> dibekacin, framycetin, gentamicin-high, habekacin, hygromycin, isepamicin,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> kanamycin-high, kanamycin/cephalexin, micronomicin, neomycin, netilmicin,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> pentisomicin, plazomicin, propikacin, ribostamycin, sisomicin,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> streptoduocin, streptomycin, streptomycin-high, and tobramycin-high</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># An Antibiogram: 2 × 5</span></span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> ℹ For aminoglycosides() using columns 'GEN' (gentamicin), 'TOB'</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> (tobramycin), 'AMK' (amikacin), and 'KAN' (kanamycin)</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># An Antibiogram (non-WISCA): 2 × 5</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> `Grupo sindrómico` Patógeno Amikacina Gentamicina Tobramicina </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">*</span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">1</span> No UCI E. coli 100% (119/119) 98% (316/323) 98% (318/325)</span>
|
||||
@ -547,29 +513,22 @@
|
||||
<span class="r-out co"><span class="r-pr">#></span> # or use it directly in R Markdown or https://quarto.org, see ?antibiogram</span>
|
||||
<span class="r-in"><span></span></span>
|
||||
<span class="r-in"><span></span></span>
|
||||
<span class="r-in"><span><span class="co"># Weighted-incidence syndromic combination antibiogram (WISCA) ---------</span></span></span>
|
||||
<span class="r-in"><span><span class="co"># WISCA antibiogram ----------------------------------------------------</span></span></span>
|
||||
<span class="r-in"><span></span></span>
|
||||
<span class="r-in"><span><span class="co"># the data set could contain a filter for e.g. respiratory specimens/ICU</span></span></span>
|
||||
<span class="r-in"><span><span class="co"># can be used for any of the above types - just add `wisca = TRUE`</span></span></span>
|
||||
<span class="r-in"><span><span class="fu">antibiogram</span><span class="op">(</span><span class="va">example_isolates</span>,</span></span>
|
||||
<span class="r-in"><span> antibiotics <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"AMC"</span>, <span class="st">"AMC+CIP"</span>, <span class="st">"TZP"</span>, <span class="st">"TZP+TOB"</span><span class="op">)</span>,</span></span>
|
||||
<span class="r-in"><span> antibiotics <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"TZP"</span>, <span class="st">"TZP+TOB"</span>, <span class="st">"TZP+GEN"</span><span class="op">)</span>,</span></span>
|
||||
<span class="r-in"><span> mo_transform <span class="op">=</span> <span class="st">"gramstain"</span>,</span></span>
|
||||
<span class="r-in"><span> minimum <span class="op">=</span> <span class="fl">10</span>, <span class="co"># this should be >=30, but now just as example</span></span></span>
|
||||
<span class="r-in"><span> syndromic_group <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/ifelse.html" class="external-link">ifelse</a></span><span class="op">(</span><span class="va">example_isolates</span><span class="op">$</span><span class="va">age</span> <span class="op">>=</span> <span class="fl">65</span> <span class="op">&</span></span></span>
|
||||
<span class="r-in"><span> <span class="va">example_isolates</span><span class="op">$</span><span class="va">gender</span> <span class="op">==</span> <span class="st">"M"</span>,</span></span>
|
||||
<span class="r-in"><span> <span class="st">"WISCA Group 1"</span>, <span class="st">"WISCA Group 2"</span></span></span>
|
||||
<span class="r-in"><span> <span class="op">)</span></span></span>
|
||||
<span class="r-in"><span> wisca <span class="op">=</span> <span class="cn">TRUE</span></span></span>
|
||||
<span class="r-in"><span><span class="op">)</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># An Antibiogram: 4 × 6</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> `Syndromic Group` Pathogen Amoxicillin/clavulani…¹ Amoxicillin/clavulan…²</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">*</span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">1</span> WISCA Group 1 Gram-negative 76% (216/285) 95% (270/284) </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">2</span> WISCA Group 2 Gram-negative 76% (336/441) 98% (432/442) </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">3</span> WISCA Group 1 Gram-positive 76% (310/406) 89% (347/392) </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">4</span> WISCA Group 2 Gram-positive 76% (556/732) 89% (617/695) </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># ℹ abbreviated names: ¹`Amoxicillin/clavulanic acid`,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># ²`Amoxicillin/clavulanic acid + Ciprofloxacin`</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># ℹ 2 more variables: `Piperacillin/tazobactam` <chr>,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># `Piperacillin/tazobactam + Tobramycin` <chr></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># An Antibiogram (WISCA / 95% CI): 2 × 4</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> Pathogen Piperacillin/tazobac…¹ Piperacillin/tazobac…² Piperacillin/tazobac…³</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">*</span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">1</span> Gram-neg… 88% (85-90%,N=641) 98% (97-99%,N=691) 98% (97-99%,N=693) </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">2</span> Gram-pos… 86% (82-89%,N=345) 97% (96-98%,N=1044) 95% (93-97%,N=550) </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># ℹ abbreviated names: ¹`Piperacillin/tazobactam`,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># ²`Piperacillin/tazobactam + Gentamicin`,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># ³`Piperacillin/tazobactam + Tobramycin`</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> # Use `plot()` or `ggplot2::autoplot()` to create a plot of this antibiogram,</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> # or use it directly in R Markdown or https://quarto.org, see ?antibiogram</span>
|
||||
<span class="r-in"><span></span></span>
|
||||
@ -580,18 +539,7 @@
|
||||
<span class="r-in"><span> antibiotics <span class="op">=</span> <span class="fu"><a href="antimicrobial_class_selectors.html">ureidopenicillins</a></span><span class="op">(</span><span class="op">)</span>,</span></span>
|
||||
<span class="r-in"><span> ab_transform <span class="op">=</span> <span class="st">"name"</span></span></span>
|
||||
<span class="r-in"><span><span class="op">)</span></span></span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> ℹ The function ureidopenicillins() should be used inside a dplyr verb</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> or data.frame call, e.g.:</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% select(ureidopenicillins())</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% select(column_a, column_b, ureidopenicillins())</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data %>% filter(any(ureidopenicillins() == "R"))</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data[, ureidopenicillins()]</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> • your_data[, c("column_a", "column_b", ureidopenicillins())]</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> </span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> Now returning a vector of all possible antimicrobials that</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> ureidopenicillins() can select.</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> <span class="warning">Warning: </span>The following antibiotics were not available and ignored: azlocillin,</span>
|
||||
<span class="r-wrn co"><span class="r-pr">#></span> mezlocillin, and piperacillin</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> ℹ For ureidopenicillins() using column 'TZP' (piperacillin/tazobactam)</span>
|
||||
<span class="r-in"><span></span></span>
|
||||
<span class="r-in"><span><span class="co"># in an Rmd file, you would just need to return `ureido` in a chunk,</span></span></span>
|
||||
<span class="r-in"><span><span class="co"># but to be explicit here:</span></span></span>
|
||||
@ -612,7 +560,8 @@
|
||||
<span class="r-in"><span></span></span>
|
||||
<span class="r-in"><span><span class="va">ab1</span> <span class="op"><-</span> <span class="fu">antibiogram</span><span class="op">(</span><span class="va">example_isolates</span>,</span></span>
|
||||
<span class="r-in"><span> antibiotics <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"AMC"</span>, <span class="st">"CIP"</span>, <span class="st">"TZP"</span>, <span class="st">"TZP+TOB"</span><span class="op">)</span>,</span></span>
|
||||
<span class="r-in"><span> mo_transform <span class="op">=</span> <span class="st">"gramstain"</span></span></span>
|
||||
<span class="r-in"><span> mo_transform <span class="op">=</span> <span class="st">"gramstain"</span>,</span></span>
|
||||
<span class="r-in"><span> wisca <span class="op">=</span> <span class="cn">TRUE</span></span></span>
|
||||
<span class="r-in"><span><span class="op">)</span></span></span>
|
||||
<span class="r-in"><span><span class="va">ab2</span> <span class="op"><-</span> <span class="fu">antibiogram</span><span class="op">(</span><span class="va">example_isolates</span>,</span></span>
|
||||
<span class="r-in"><span> antibiotics <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"AMC"</span>, <span class="st">"CIP"</span>, <span class="st">"TZP"</span>, <span class="st">"TZP+TOB"</span><span class="op">)</span>,</span></span>
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -17,7 +17,7 @@ my_data_with_all_these_columns %&gt;%
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -68,28 +68,14 @@ my_data_with_all_these_columns %&gt;%
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">amr_class</span><span class="op">(</span></span>
|
||||
<span> <span class="va">amr_class</span>,</span>
|
||||
<span> only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_treatable <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> return_all <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">amr_class</span><span class="op">(</span><span class="va">amr_class</span>, only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, only_treatable <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> return_all <span class="op">=</span> <span class="cn">TRUE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">amr_selector</span><span class="op">(</span></span>
|
||||
<span> <span class="va">filter</span>,</span>
|
||||
<span> only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_treatable <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> return_all <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">amr_selector</span><span class="op">(</span><span class="va">filter</span>, only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, only_treatable <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> return_all <span class="op">=</span> <span class="cn">TRUE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">aminoglycosides</span><span class="op">(</span></span>
|
||||
<span> only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_treatable <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> return_all <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">aminoglycosides</span><span class="op">(</span>only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, only_treatable <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> return_all <span class="op">=</span> <span class="cn">TRUE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">aminopenicillins</span><span class="op">(</span>only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, return_all <span class="op">=</span> <span class="cn">TRUE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
@ -97,21 +83,14 @@ my_data_with_all_these_columns %&gt;%
|
||||
<span></span>
|
||||
<span><span class="fu">antimycobacterials</span><span class="op">(</span>only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, return_all <span class="op">=</span> <span class="cn">TRUE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">betalactams</span><span class="op">(</span></span>
|
||||
<span> only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_treatable <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> return_all <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">betalactams</span><span class="op">(</span>only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, only_treatable <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> return_all <span class="op">=</span> <span class="cn">TRUE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">betalactams_with_inhibitor</span><span class="op">(</span>only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, return_all <span class="op">=</span> <span class="cn">TRUE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span><span class="fu">betalactams_with_inhibitor</span><span class="op">(</span>only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, return_all <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">carbapenems</span><span class="op">(</span></span>
|
||||
<span> only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_treatable <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> return_all <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">carbapenems</span><span class="op">(</span>only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, only_treatable <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> return_all <span class="op">=</span> <span class="cn">TRUE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">cephalosporins</span><span class="op">(</span>only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, return_all <span class="op">=</span> <span class="cn">TRUE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
@ -129,12 +108,8 @@ my_data_with_all_these_columns %&gt;%
|
||||
<span></span>
|
||||
<span><span class="fu">glycopeptides</span><span class="op">(</span>only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, return_all <span class="op">=</span> <span class="cn">TRUE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">lincosamides</span><span class="op">(</span></span>
|
||||
<span> only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_treatable <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> return_all <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">lincosamides</span><span class="op">(</span>only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, only_treatable <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> return_all <span class="op">=</span> <span class="cn">TRUE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">lipoglycopeptides</span><span class="op">(</span>only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, return_all <span class="op">=</span> <span class="cn">TRUE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
@ -148,12 +123,8 @@ my_data_with_all_these_columns %&gt;%
|
||||
<span></span>
|
||||
<span><span class="fu">phenicols</span><span class="op">(</span>only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, return_all <span class="op">=</span> <span class="cn">TRUE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">polymyxins</span><span class="op">(</span></span>
|
||||
<span> only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_treatable <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> return_all <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">polymyxins</span><span class="op">(</span>only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, only_treatable <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> return_all <span class="op">=</span> <span class="cn">TRUE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">quinolones</span><span class="op">(</span>only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, return_all <span class="op">=</span> <span class="cn">TRUE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
@ -171,12 +142,8 @@ my_data_with_all_these_columns %&gt;%
|
||||
<span></span>
|
||||
<span><span class="fu">administrable_iv</span><span class="op">(</span>only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, return_all <span class="op">=</span> <span class="cn">TRUE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">not_intrinsic_resistant</span><span class="op">(</span></span>
|
||||
<span> only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> col_mo <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> version_expertrules <span class="op">=</span> <span class="fl">3.3</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span></code></pre></div>
|
||||
<span><span class="fu">not_intrinsic_resistant</span><span class="op">(</span>only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, col_mo <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> version_expertrules <span class="op">=</span> <span class="fl">3.3</span>, <span class="va">...</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -54,20 +54,14 @@
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">as.mo</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> Becker <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> Lancefield <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">as.mo</span><span class="op">(</span><span class="va">x</span>, Becker <span class="op">=</span> <span class="cn">FALSE</span>, Lancefield <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> minimum_matching_score <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> reference_df <span class="op">=</span> <span class="fu"><a href="mo_source.html">get_mo_source</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> ignore_pattern <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_ignore_pattern"</span>, <span class="cn">NULL</span><span class="op">)</span>,</span>
|
||||
<span> cleaning_regex <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_cleaning_regex"</span>, <span class="fu">mo_cleaning_regex</span><span class="op">(</span><span class="op">)</span><span class="op">)</span>,</span>
|
||||
<span> only_fungi <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_only_fungi"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> info <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/interactive.html" class="external-link">interactive</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>, info <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/interactive.html" class="external-link">interactive</a></span><span class="op">(</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">is.mo</span><span class="op">(</span><span class="va">x</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
|
||||
@ -21,7 +21,7 @@ All breakpoints used for interpretation are available in our clinical_breakpoint
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -81,67 +81,38 @@ All breakpoints used for interpretation are available in our clinical_breakpoint
|
||||
<span><span class="fu">is_sir_eligible</span><span class="op">(</span><span class="va">x</span>, threshold <span class="op">=</span> <span class="fl">0.05</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="co"># Default S3 method</span></span>
|
||||
<span><span class="fu">as.sir</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> S <span class="op">=</span> <span class="st">"^(S|U)+$"</span>,</span>
|
||||
<span> I <span class="op">=</span> <span class="st">"^(I)+$"</span>,</span>
|
||||
<span> R <span class="op">=</span> <span class="st">"^(R)+$"</span>,</span>
|
||||
<span> NI <span class="op">=</span> <span class="st">"^(N|NI|V)+$"</span>,</span>
|
||||
<span> SDD <span class="op">=</span> <span class="st">"^(SDD|D|H)+$"</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">as.sir</span><span class="op">(</span><span class="va">x</span>, S <span class="op">=</span> <span class="st">"^(S|U)+$"</span>, I <span class="op">=</span> <span class="st">"^(I)+$"</span>, R <span class="op">=</span> <span class="st">"^(R)+$"</span>,</span>
|
||||
<span> NI <span class="op">=</span> <span class="st">"^(N|NI|V)+$"</span>, SDD <span class="op">=</span> <span class="st">"^(SDD|D|H)+$"</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="co"># S3 method for class 'mic'</span></span>
|
||||
<span><span class="fu">as.sir</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> mo <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> ab <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/deparse.html" class="external-link">deparse</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/substitute.html" class="external-link">substitute</a></span><span class="op">(</span><span class="va">x</span><span class="op">)</span><span class="op">)</span>,</span>
|
||||
<span> guideline <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_guideline"</span>, <span class="st">"EUCAST"</span><span class="op">)</span>,</span>
|
||||
<span> uti <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> conserve_capped_values <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> add_intrinsic_resistance <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span><span class="fu">as.sir</span><span class="op">(</span><span class="va">x</span>, mo <span class="op">=</span> <span class="cn">NULL</span>, ab <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/deparse.html" class="external-link">deparse</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/substitute.html" class="external-link">substitute</a></span><span class="op">(</span><span class="va">x</span><span class="op">)</span><span class="op">)</span>,</span>
|
||||
<span> guideline <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_guideline"</span>, <span class="st">"EUCAST"</span><span class="op">)</span>, uti <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> conserve_capped_values <span class="op">=</span> <span class="cn">FALSE</span>, add_intrinsic_resistance <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> reference_data <span class="op">=</span> <span class="fu">AMR</span><span class="fu">::</span><span class="va"><a href="clinical_breakpoints.html">clinical_breakpoints</a></span>,</span>
|
||||
<span> include_screening <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_include_screening"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> include_PKPD <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_include_PKPD"</span>, <span class="cn">TRUE</span><span class="op">)</span>,</span>
|
||||
<span> breakpoint_type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_breakpoint_type"</span>, <span class="st">"human"</span><span class="op">)</span>,</span>
|
||||
<span> host <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> verbose <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span> breakpoint_type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_breakpoint_type"</span>, <span class="st">"human"</span><span class="op">)</span>, host <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> verbose <span class="op">=</span> <span class="cn">FALSE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="co"># S3 method for class 'disk'</span></span>
|
||||
<span><span class="fu">as.sir</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> mo <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> ab <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/deparse.html" class="external-link">deparse</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/substitute.html" class="external-link">substitute</a></span><span class="op">(</span><span class="va">x</span><span class="op">)</span><span class="op">)</span>,</span>
|
||||
<span> guideline <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_guideline"</span>, <span class="st">"EUCAST"</span><span class="op">)</span>,</span>
|
||||
<span> uti <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span><span class="fu">as.sir</span><span class="op">(</span><span class="va">x</span>, mo <span class="op">=</span> <span class="cn">NULL</span>, ab <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/deparse.html" class="external-link">deparse</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/substitute.html" class="external-link">substitute</a></span><span class="op">(</span><span class="va">x</span><span class="op">)</span><span class="op">)</span>,</span>
|
||||
<span> guideline <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_guideline"</span>, <span class="st">"EUCAST"</span><span class="op">)</span>, uti <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> add_intrinsic_resistance <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> reference_data <span class="op">=</span> <span class="fu">AMR</span><span class="fu">::</span><span class="va"><a href="clinical_breakpoints.html">clinical_breakpoints</a></span>,</span>
|
||||
<span> include_screening <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_include_screening"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> include_PKPD <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_include_PKPD"</span>, <span class="cn">TRUE</span><span class="op">)</span>,</span>
|
||||
<span> breakpoint_type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_breakpoint_type"</span>, <span class="st">"human"</span><span class="op">)</span>,</span>
|
||||
<span> host <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> verbose <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span> breakpoint_type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_breakpoint_type"</span>, <span class="st">"human"</span><span class="op">)</span>, host <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> verbose <span class="op">=</span> <span class="cn">FALSE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="co"># S3 method for class 'data.frame'</span></span>
|
||||
<span><span class="fu">as.sir</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> <span class="va">...</span>,</span>
|
||||
<span> col_mo <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> guideline <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_guideline"</span>, <span class="st">"EUCAST"</span><span class="op">)</span>,</span>
|
||||
<span> uti <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> conserve_capped_values <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> add_intrinsic_resistance <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span><span class="fu">as.sir</span><span class="op">(</span><span class="va">x</span>, <span class="va">...</span>, col_mo <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> guideline <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_guideline"</span>, <span class="st">"EUCAST"</span><span class="op">)</span>, uti <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> conserve_capped_values <span class="op">=</span> <span class="cn">FALSE</span>, add_intrinsic_resistance <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> reference_data <span class="op">=</span> <span class="fu">AMR</span><span class="fu">::</span><span class="va"><a href="clinical_breakpoints.html">clinical_breakpoints</a></span>,</span>
|
||||
<span> include_screening <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_include_screening"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> include_PKPD <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_include_PKPD"</span>, <span class="cn">TRUE</span><span class="op">)</span>,</span>
|
||||
<span> breakpoint_type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_breakpoint_type"</span>, <span class="st">"human"</span><span class="op">)</span>,</span>
|
||||
<span> host <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> verbose <span class="op">=</span> <span class="cn">FALSE</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span> breakpoint_type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_breakpoint_type"</span>, <span class="st">"human"</span><span class="op">)</span>, host <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> verbose <span class="op">=</span> <span class="cn">FALSE</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">sir_interpretation_history</span><span class="op">(</span>clean <span class="op">=</span> <span class="cn">FALSE</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
@ -786,16 +757,16 @@ A microorganism is categorised as "Resistant" when there is a high likelihood of
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># A tibble: 57 × 16</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> datetime index ab_given mo_given host_given ab mo </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">*</span> <span style="color: #949494; font-style: italic;"><dttm></span> <span style="color: #949494; font-style: italic;"><int></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><ab></span> <span style="color: #949494; font-style: italic;"><mo></span> </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 1</span> 2025-01-17 <span style="color: #949494;">11:16:11</span> 4 AMX B_STRPT… human AMX B_STRPT_PNMN </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 2</span> 2025-01-17 <span style="color: #949494;">11:16:18</span> 4 genta Escheri… human GEN B_[ORD]_ENTRBCTR</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 3</span> 2025-01-17 <span style="color: #949494;">11:16:18</span> 4 genta Escheri… human GEN B_[ORD]_ENTRBCTR</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 4</span> 2025-01-17 <span style="color: #949494;">11:16:19</span> 4 genta Escheri… cattle GEN B_ESCHR_COLI </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 5</span> 2025-01-17 <span style="color: #949494;">11:16:19</span> 4 genta Escheri… cattle GEN B_ESCHR_COLI </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 6</span> 2025-01-17 <span style="color: #949494;">11:16:11</span> 3 AMX B_STRPT… human AMX B_STRPT_PNMN </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 7</span> 2025-01-17 <span style="color: #949494;">11:16:18</span> 3 tobra Escheri… human TOB B_[ORD]_ENTRBCTR</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 8</span> 2025-01-17 <span style="color: #949494;">11:16:18</span> 3 tobra Escheri… human TOB B_[ORD]_ENTRBCTR</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 9</span> 2025-01-17 <span style="color: #949494;">11:16:19</span> 3 tobra Escheri… horses TOB B_ESCHR_COLI </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">10</span> 2025-01-17 <span style="color: #949494;">11:16:19</span> 3 tobra Escheri… horses TOB B_ESCHR_COLI </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 1</span> 2025-01-27 <span style="color: #949494;">21:13:26</span> 4 AMX B_STRPT… human AMX B_STRPT_PNMN </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 2</span> 2025-01-27 <span style="color: #949494;">21:13:33</span> 4 genta Escheri… human GEN B_[ORD]_ENTRBCTR</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 3</span> 2025-01-27 <span style="color: #949494;">21:13:34</span> 4 genta Escheri… human GEN B_[ORD]_ENTRBCTR</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 4</span> 2025-01-27 <span style="color: #949494;">21:13:34</span> 4 genta Escheri… cattle GEN B_ESCHR_COLI </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 5</span> 2025-01-27 <span style="color: #949494;">21:13:34</span> 4 genta Escheri… cattle GEN B_ESCHR_COLI </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 6</span> 2025-01-27 <span style="color: #949494;">21:13:26</span> 3 AMX B_STRPT… human AMX B_STRPT_PNMN </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 7</span> 2025-01-27 <span style="color: #949494;">21:13:33</span> 3 tobra Escheri… human TOB B_[ORD]_ENTRBCTR</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 8</span> 2025-01-27 <span style="color: #949494;">21:13:33</span> 3 tobra Escheri… human TOB B_[ORD]_ENTRBCTR</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 9</span> 2025-01-27 <span style="color: #949494;">21:13:34</span> 3 tobra Escheri… horses TOB B_ESCHR_COLI </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">10</span> 2025-01-27 <span style="color: #949494;">21:13:34</span> 3 tobra Escheri… horses TOB B_ESCHR_COLI </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># ℹ 47 more rows</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># ℹ 9 more variables: host <chr>, method <chr>, input <dbl>, outcome <sir>,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># notes <chr>, guideline <chr>, ref_table <chr>, uti <lgl>,</span></span>
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -54,13 +54,9 @@
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">atc_online_property</span><span class="op">(</span></span>
|
||||
<span> <span class="va">atc_code</span>,</span>
|
||||
<span> <span class="va">property</span>,</span>
|
||||
<span> administration <span class="op">=</span> <span class="st">"O"</span>,</span>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">atc_online_property</span><span class="op">(</span><span class="va">atc_code</span>, <span class="va">property</span>, administration <span class="op">=</span> <span class="st">"O"</span>,</span>
|
||||
<span> url <span class="op">=</span> <span class="st">"https://atcddd.fhi.no/atc_ddd_index/?code=%s&showdescription=no"</span>,</span>
|
||||
<span> url_vet <span class="op">=</span> <span class="st">"https://atcddd.fhi.no/atcvet/atcvet_index/?code=%s&showdescription=no"</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span> url_vet <span class="op">=</span> <span class="st">"https://atcddd.fhi.no/atcvet/atcvet_index/?code=%s&showdescription=no"</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">atc_online_groups</span><span class="op">(</span><span class="va">atc_code</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -54,15 +54,9 @@
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">av_from_text</span><span class="op">(</span></span>
|
||||
<span> <span class="va">text</span>,</span>
|
||||
<span> type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"drug"</span>, <span class="st">"dose"</span>, <span class="st">"administration"</span><span class="op">)</span>,</span>
|
||||
<span> collapse <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> translate_av <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> thorough_search <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> info <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/interactive.html" class="external-link">interactive</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span></code></pre></div>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">av_from_text</span><span class="op">(</span><span class="va">text</span>, type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"drug"</span>, <span class="st">"dose"</span>, <span class="st">"administration"</span><span class="op">)</span>,</span>
|
||||
<span> collapse <span class="op">=</span> <span class="cn">NULL</span>, translate_av <span class="op">=</span> <span class="cn">FALSE</span>, thorough_search <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> info <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/interactive.html" class="external-link">interactive</a></span><span class="op">(</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -54,21 +54,15 @@
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">bug_drug_combinations</span><span class="op">(</span><span class="va">x</span>, col_mo <span class="op">=</span> <span class="cn">NULL</span>, FUN <span class="op">=</span> <span class="va">mo_shortname</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">bug_drug_combinations</span><span class="op">(</span><span class="va">x</span>, col_mo <span class="op">=</span> <span class="cn">NULL</span>, FUN <span class="op">=</span> <span class="va">mo_shortname</span>,</span>
|
||||
<span> include_n_rows <span class="op">=</span> <span class="cn">FALSE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="co"># S3 method for class 'bug_drug_combinations'</span></span>
|
||||
<span><span class="fu"><a href="https://rdrr.io/r/base/format.html" class="external-link">format</a></span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> translate_ab <span class="op">=</span> <span class="st">"name (ab, atc)"</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> minimum <span class="op">=</span> <span class="fl">30</span>,</span>
|
||||
<span> combine_SI <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> add_ab_group <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> remove_intrinsic_resistant <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> decimal.mark <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"OutDec"</span><span class="op">)</span>,</span>
|
||||
<span> big.mark <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/ifelse.html" class="external-link">ifelse</a></span><span class="op">(</span><span class="va">decimal.mark</span> <span class="op">==</span> <span class="st">","</span>, <span class="st">"."</span>, <span class="st">","</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span></code></pre></div>
|
||||
<span><span class="fu"><a href="https://rdrr.io/r/base/format.html" class="external-link">format</a></span><span class="op">(</span><span class="va">x</span>, translate_ab <span class="op">=</span> <span class="st">"name (ab, atc)"</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>, minimum <span class="op">=</span> <span class="fl">30</span>, combine_SI <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> add_ab_group <span class="op">=</span> <span class="cn">TRUE</span>, remove_intrinsic_resistant <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> decimal.mark <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"OutDec"</span><span class="op">)</span>, big.mark <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/ifelse.html" class="external-link">ifelse</a></span><span class="op">(</span><span class="va">decimal.mark</span> <span class="op">==</span></span>
|
||||
<span> <span class="st">","</span>, <span class="st">"."</span>, <span class="st">","</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
@ -87,6 +81,10 @@
|
||||
<dd><p>the function to call on the <code>mo</code> column to transform the microorganism codes - the default is <code><a href="mo_property.html">mo_shortname()</a></code></p></dd>
|
||||
|
||||
|
||||
<dt id="arg-include-n-rows">include_n_rows<a class="anchor" aria-label="anchor" href="#arg-include-n-rows"></a></dt>
|
||||
<dd><p>a <a href="https://rdrr.io/r/base/logical.html" class="external-link">logical</a> to indicate if the total number of rows must be included in the output</p></dd>
|
||||
|
||||
|
||||
<dt id="arg--">...<a class="anchor" aria-label="anchor" href="#arg--"></a></dt>
|
||||
<dd><p>arguments passed on to <code>FUN</code></p></dd>
|
||||
|
||||
|
||||
@ -21,7 +21,7 @@ Use as.sir() to transform MICs or disks measurements to SIR values."><meta prope
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -9,7 +9,7 @@ count_resistant() should be used to count resistant isolates, count_susceptible(
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -75,12 +75,8 @@ count_resistant() should be used to count resistant isolates, count_susceptible(
|
||||
<span></span>
|
||||
<span><span class="fu">n_sir</span><span class="op">(</span><span class="va">...</span>, only_all_tested <span class="op">=</span> <span class="cn">FALSE</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">count_df</span><span class="op">(</span></span>
|
||||
<span> <span class="va">data</span>,</span>
|
||||
<span> translate_ab <span class="op">=</span> <span class="st">"name"</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> combine_SI <span class="op">=</span> <span class="cn">TRUE</span></span>
|
||||
<span><span class="op">)</span></span></code></pre></div>
|
||||
<span><span class="fu">count_df</span><span class="op">(</span><span class="va">data</span>, translate_ab <span class="op">=</span> <span class="st">"name"</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> combine_SI <span class="op">=</span> <span class="cn">TRUE</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -9,7 +9,7 @@ To improve the interpretation of the antibiogram before EUCAST rules are applied
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -57,19 +57,11 @@ To improve the interpretation of the antibiogram before EUCAST rules are applied
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">eucast_rules</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> col_mo <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> info <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/interactive.html" class="external-link">interactive</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">eucast_rules</span><span class="op">(</span><span class="va">x</span>, col_mo <span class="op">=</span> <span class="cn">NULL</span>, info <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/interactive.html" class="external-link">interactive</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> rules <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_eucastrules"</span>, default <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"breakpoints"</span>, <span class="st">"expert"</span><span class="op">)</span><span class="op">)</span>,</span>
|
||||
<span> verbose <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> version_breakpoints <span class="op">=</span> <span class="fl">12</span>,</span>
|
||||
<span> version_expertrules <span class="op">=</span> <span class="fl">3.3</span>,</span>
|
||||
<span> ampc_cephalosporin_resistance <span class="op">=</span> <span class="cn">NA</span>,</span>
|
||||
<span> only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> custom_rules <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span> verbose <span class="op">=</span> <span class="cn">FALSE</span>, version_breakpoints <span class="op">=</span> <span class="fl">12</span>, version_expertrules <span class="op">=</span> <span class="fl">3.3</span>,</span>
|
||||
<span> ampc_cephalosporin_resistance <span class="op">=</span> <span class="cn">NA</span>, only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> custom_rules <span class="op">=</span> <span class="cn">NULL</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">eucast_dosage</span><span class="op">(</span><span class="va">ab</span>, administration <span class="op">=</span> <span class="st">"iv"</span>, version_breakpoints <span class="op">=</span> <span class="fl">12</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -54,13 +54,9 @@
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">export_ncbi_biosample</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> filename <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/paste.html" class="external-link">paste0</a></span><span class="op">(</span><span class="st">"biosample_"</span>, <span class="fu"><a href="https://rdrr.io/r/base/format.html" class="external-link">format</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/Sys.time.html" class="external-link">Sys.time</a></span><span class="op">(</span><span class="op">)</span>, <span class="st">"%Y-%m-%d-%H%M%S"</span><span class="op">)</span>, <span class="st">".xlsx"</span><span class="op">)</span>,</span>
|
||||
<span> type <span class="op">=</span> <span class="st">"pathogen MIC"</span>,</span>
|
||||
<span> columns <span class="op">=</span> <span class="fu"><a href="https://tidyselect.r-lib.org/reference/where.html" class="external-link">where</a></span><span class="op">(</span><span class="va">is.mic</span><span class="op">)</span>,</span>
|
||||
<span> save_as_xlsx <span class="op">=</span> <span class="cn">TRUE</span></span>
|
||||
<span><span class="op">)</span></span></code></pre></div>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">export_ncbi_biosample</span><span class="op">(</span><span class="va">x</span>, filename <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/paste.html" class="external-link">paste0</a></span><span class="op">(</span><span class="st">"biosample_"</span>, <span class="fu"><a href="https://rdrr.io/r/base/format.html" class="external-link">format</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/Sys.time.html" class="external-link">Sys.time</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> <span class="st">"%Y-%m-%d-%H%M%S"</span><span class="op">)</span>, <span class="st">".xlsx"</span><span class="op">)</span>, type <span class="op">=</span> <span class="st">"pathogen MIC"</span>,</span>
|
||||
<span> columns <span class="op">=</span> <span class="fu"><a href="https://tidyselect.r-lib.org/reference/where.html" class="external-link">where</a></span><span class="op">(</span><span class="va">is.mic</span><span class="op">)</span>, save_as_xlsx <span class="op">=</span> <span class="cn">TRUE</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
|
||||
@ -9,7 +9,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -57,38 +57,18 @@
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">first_isolate</span><span class="op">(</span></span>
|
||||
<span> x <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> col_date <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> col_patient_id <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> col_mo <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> col_testcode <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> col_specimen <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> col_icu <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> col_keyantimicrobials <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> episode_days <span class="op">=</span> <span class="fl">365</span>,</span>
|
||||
<span> testcodes_exclude <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> icu_exclude <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> specimen_group <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> type <span class="op">=</span> <span class="st">"points"</span>,</span>
|
||||
<span> method <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"phenotype-based"</span>, <span class="st">"episode-based"</span>, <span class="st">"patient-based"</span>, <span class="st">"isolate-based"</span><span class="op">)</span>,</span>
|
||||
<span> ignore_I <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> points_threshold <span class="op">=</span> <span class="fl">2</span>,</span>
|
||||
<span> info <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/interactive.html" class="external-link">interactive</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> include_unknown <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> include_untested_sir <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">first_isolate</span><span class="op">(</span>x <span class="op">=</span> <span class="cn">NULL</span>, col_date <span class="op">=</span> <span class="cn">NULL</span>, col_patient_id <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> col_mo <span class="op">=</span> <span class="cn">NULL</span>, col_testcode <span class="op">=</span> <span class="cn">NULL</span>, col_specimen <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> col_icu <span class="op">=</span> <span class="cn">NULL</span>, col_keyantimicrobials <span class="op">=</span> <span class="cn">NULL</span>, episode_days <span class="op">=</span> <span class="fl">365</span>,</span>
|
||||
<span> testcodes_exclude <span class="op">=</span> <span class="cn">NULL</span>, icu_exclude <span class="op">=</span> <span class="cn">FALSE</span>, specimen_group <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> type <span class="op">=</span> <span class="st">"points"</span>, method <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"phenotype-based"</span>, <span class="st">"episode-based"</span>,</span>
|
||||
<span> <span class="st">"patient-based"</span>, <span class="st">"isolate-based"</span><span class="op">)</span>, ignore_I <span class="op">=</span> <span class="cn">TRUE</span>, points_threshold <span class="op">=</span> <span class="fl">2</span>,</span>
|
||||
<span> info <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/interactive.html" class="external-link">interactive</a></span><span class="op">(</span><span class="op">)</span>, include_unknown <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> include_untested_sir <span class="op">=</span> <span class="cn">TRUE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">filter_first_isolate</span><span class="op">(</span></span>
|
||||
<span> x <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> col_date <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> col_patient_id <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> col_mo <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> episode_days <span class="op">=</span> <span class="fl">365</span>,</span>
|
||||
<span> method <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"phenotype-based"</span>, <span class="st">"episode-based"</span>, <span class="st">"patient-based"</span>, <span class="st">"isolate-based"</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span></code></pre></div>
|
||||
<span><span class="fu">filter_first_isolate</span><span class="op">(</span>x <span class="op">=</span> <span class="cn">NULL</span>, col_date <span class="op">=</span> <span class="cn">NULL</span>, col_patient_id <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> col_mo <span class="op">=</span> <span class="cn">NULL</span>, episode_days <span class="op">=</span> <span class="fl">365</span>, method <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"phenotype-based"</span>,</span>
|
||||
<span> <span class="st">"episode-based"</span>, <span class="st">"patient-based"</span>, <span class="st">"isolate-based"</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
@ -186,15 +166,16 @@
|
||||
</div>
|
||||
<div class="section level2">
|
||||
<h2 id="details">Details<a class="anchor" aria-label="anchor" href="#details"></a></h2>
|
||||
<p>To conduct epidemiological analyses on antimicrobial resistance data, only so-called first isolates should be included to prevent overestimation and underestimation of antimicrobial resistance. Different methods can be used to do so, see below.</p>
|
||||
<p>The methodology implemented in these functions is based on the research overview by Hindler <em>et al.</em> (2007, <a href="https://doi.org/10.1086/511864" class="external-link">doi:10.1086/511864</a>
|
||||
) and the recommendations outlined in the <a href="https://clsi.org/standards/products/microbiology/documents/m39" class="external-link">CLSI Guideline M39</a>.
|
||||
To conduct epidemiological analyses on antimicrobial resistance data, only so-called first isolates should be included to prevent overestimation and underestimation of antimicrobial resistance. Different methods can be used to do so, see below.</p>
|
||||
<p>These functions are context-aware. This means that the <code>x</code> argument can be left blank if used inside a <a href="https://rdrr.io/r/base/data.frame.html" class="external-link">data.frame</a> call, see <em>Examples</em>.</p>
|
||||
<p>The <code>first_isolate()</code> function is a wrapper around the <code><a href="get_episode.html">is_new_episode()</a></code> function, but more efficient for data sets containing microorganism codes or names.</p>
|
||||
<p>All isolates with a microbial ID of <code>NA</code> will be excluded as first isolate.</p><div class="section">
|
||||
<h3 id="different-methods">Different methods<a class="anchor" aria-label="anchor" href="#different-methods"></a></h3>
|
||||
|
||||
|
||||
<p>According to Hindler <em>et al.</em> (2007, <a href="https://doi.org/10.1086/511864" class="external-link">doi:10.1086/511864</a>
|
||||
), there are different methods (algorithms) to select first isolates with increasing reliability: isolate-based, patient-based, episode-based and phenotype-based. All methods select on a combination of the taxonomic genus and species (not subspecies).</p>
|
||||
<p>According to previously-mentioned sources, there are different methods (algorithms) to select first isolates with increasing reliability: isolate-based, patient-based, episode-based and phenotype-based. All methods select on a combination of the taxonomic genus and species (not subspecies).</p>
|
||||
<p>All mentioned methods are covered in the <code>first_isolate()</code> function:</p><table class="table table"><tr><td><strong>Method</strong></td><td><strong>Function to apply</strong></td></tr><tr><td><strong>Isolate-based</strong></td><td><code>first_isolate(x, method = "isolate-based")</code></td></tr><tr><td><em>(= all isolates)</em></td><td></td></tr><tr><td></td><td></td></tr><tr><td></td><td></td></tr><tr><td><strong>Patient-based</strong></td><td><code>first_isolate(x, method = "patient-based")</code></td></tr><tr><td><em>(= first isolate per patient)</em></td><td></td></tr><tr><td></td><td></td></tr><tr><td></td><td></td></tr><tr><td><strong>Episode-based</strong></td><td><code>first_isolate(x, method = "episode-based")</code>, or:</td></tr><tr><td><em>(= first isolate per episode)</em></td><td></td></tr><tr><td>- 7-Day interval from initial isolate</td><td>- <code>first_isolate(x, method = "e", episode_days = 7)</code></td></tr><tr><td>- 30-Day interval from initial isolate</td><td>- <code>first_isolate(x, method = "e", episode_days = 30)</code></td></tr><tr><td></td><td></td></tr><tr><td></td><td></td></tr><tr><td><strong>Phenotype-based</strong></td><td><code>first_isolate(x, method = "phenotype-based")</code>, or:</td></tr><tr><td><em>(= first isolate per phenotype)</em></td><td></td></tr><tr><td>- Major difference in any antimicrobial result</td><td>- <code>first_isolate(x, type = "points")</code></td></tr><tr><td>- Any difference in key antimicrobial results</td><td>- <code>first_isolate(x, type = "keyantimicrobials")</code></td></tr></table></div>
|
||||
|
||||
<div class="section">
|
||||
@ -208,14 +189,14 @@
|
||||
<h3 id="patient-based">Patient-based<a class="anchor" aria-label="anchor" href="#patient-based"></a></h3>
|
||||
|
||||
|
||||
<p>To include every genus-species combination per patient once, set the <code>episode_days</code> to <code>Inf</code>. Although often inappropriate, this method makes sure that no duplicate isolates are selected from the same patient. In a large longitudinal data set, this could mean that isolates are <em>excluded</em> that were found years after the initial isolate.</p>
|
||||
<p>To include every genus-species combination per patient once, set the <code>episode_days</code> to <code>Inf</code>. This method makes sure that no duplicate isolates are selected from the same patient. This method is preferred to e.g. identify the first MRSA finding of each patient to determine the incidence. Conversely, in a large longitudinal data set, this could mean that isolates are <em>excluded</em> that were found years after the initial isolate.</p>
|
||||
</div>
|
||||
|
||||
<div class="section">
|
||||
<h3 id="episode-based">Episode-based<a class="anchor" aria-label="anchor" href="#episode-based"></a></h3>
|
||||
|
||||
|
||||
<p>To include every genus-species combination per patient episode once, set the <code>episode_days</code> to a sensible number of days. Depending on the type of analysis, this could be 14, 30, 60 or 365. Short episodes are common for analysing specific hospital or ward data, long episodes are common for analysing regional and national data.</p>
|
||||
<p>To include every genus-species combination per patient episode once, set the <code>episode_days</code> to a sensible number of days. Depending on the type of analysis, this could be 14, 30, 60 or 365. Short episodes are common for analysing specific hospital or ward data or ICU cases, long episodes are common for analysing regional and national data.</p>
|
||||
<p>This is the most common method to correct for duplicate isolates. Patients are categorised into episodes based on their ID and dates (e.g., the date of specimen receipt or laboratory result). While this is a common method, it does not take into account antimicrobial test results. This means that e.g. a methicillin-resistant <em>Staphylococcus aureus</em> (MRSA) isolate cannot be differentiated from a wildtype <em>Staphylococcus aureus</em> isolate.</p>
|
||||
</div>
|
||||
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -153,27 +153,28 @@
|
||||
<span class="r-in"><span><span class="va">df</span> <span class="op"><-</span> <span class="va">example_isolates</span><span class="op">[</span><span class="fu"><a href="https://rdrr.io/r/base/sample.html" class="external-link">sample</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/seq.html" class="external-link">seq_len</a></span><span class="op">(</span><span class="fl">2000</span><span class="op">)</span>, size <span class="op">=</span> <span class="fl">100</span><span class="op">)</span>, <span class="op">]</span></span></span>
|
||||
<span class="r-in"><span></span></span>
|
||||
<span class="r-in"><span><span class="fu">get_episode</span><span class="op">(</span><span class="va">df</span><span class="op">$</span><span class="va">date</span>, episode_days <span class="op">=</span> <span class="fl">60</span><span class="op">)</span> <span class="co"># indices</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 41 37 28 7 11 41 48 47 3 1 32 8 1 1 28 15 28 31 25 27 20 38 33 4 45</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [26] 23 15 8 13 44 34 16 42 14 6 4 47 4 10 15 41 16 24 2 14 26 21 20 19 35</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [51] 18 17 11 43 17 15 17 2 22 21 20 10 31 9 30 12 22 18 47 11 30 16 5 46 36</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [76] 21 20 14 29 18 39 28 7 27 33 10 6 26 22 25 47 40 39 14 14 39 1 34 42 31</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 4 33 6 2 25 6 21 16 50 24 16 7 22 41 47 35 6 1 20 27 16 25 20 4 38</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [26] 16 31 43 40 10 8 24 9 19 8 28 14 32 11 39 36 49 5 8 33 17 9 34 4 19</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [51] 18 29 7 50 45 10 30 13 10 13 27 23 43 26 15 14 43 49 37 39 10 26 4 15 28</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [76] 48 38 19 50 26 32 3 42 24 28 21 37 7 42 44 41 4 3 46 5 1 49 48 34 12</span>
|
||||
<span class="r-in"><span><span class="fu">is_new_episode</span><span class="op">(</span><span class="va">df</span><span class="op">$</span><span class="va">date</span>, episode_days <span class="op">=</span> <span class="fl">60</span><span class="op">)</span> <span class="co"># TRUE/FALSE</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] TRUE TRUE TRUE TRUE TRUE FALSE TRUE TRUE TRUE TRUE TRUE TRUE</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [13] FALSE FALSE FALSE TRUE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [25] TRUE TRUE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE FALSE</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [37] FALSE FALSE TRUE FALSE FALSE FALSE TRUE TRUE FALSE TRUE TRUE FALSE</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [49] TRUE TRUE TRUE TRUE FALSE TRUE FALSE FALSE FALSE FALSE TRUE FALSE</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [61] FALSE FALSE FALSE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [73] TRUE TRUE TRUE FALSE FALSE FALSE TRUE FALSE TRUE FALSE FALSE FALSE</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [85] FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE FALSE</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [97] FALSE FALSE FALSE FALSE</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] TRUE TRUE TRUE TRUE TRUE FALSE TRUE TRUE TRUE TRUE FALSE TRUE</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [13] TRUE TRUE TRUE TRUE FALSE TRUE TRUE TRUE FALSE FALSE FALSE FALSE</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [25] TRUE FALSE TRUE TRUE TRUE TRUE TRUE FALSE TRUE TRUE FALSE TRUE</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [37] TRUE TRUE TRUE TRUE TRUE TRUE TRUE FALSE FALSE TRUE FALSE TRUE</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [49] FALSE FALSE TRUE TRUE FALSE FALSE TRUE FALSE TRUE TRUE FALSE FALSE</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [61] FALSE TRUE FALSE TRUE TRUE FALSE FALSE FALSE TRUE FALSE FALSE FALSE</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [73] FALSE FALSE FALSE TRUE FALSE FALSE FALSE FALSE FALSE TRUE TRUE FALSE</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [85] FALSE FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE TRUE FALSE FALSE</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [97] FALSE FALSE FALSE TRUE</span>
|
||||
<span class="r-in"><span></span></span>
|
||||
<span class="r-in"><span><span class="co"># filter on results from the third 60-day episode only, using base R</span></span></span>
|
||||
<span class="r-in"><span><span class="va">df</span><span class="op">[</span><span class="fu"><a href="https://rdrr.io/r/base/which.html" class="external-link">which</a></span><span class="op">(</span><span class="fu">get_episode</span><span class="op">(</span><span class="va">df</span><span class="op">$</span><span class="va">date</span>, <span class="fl">60</span><span class="op">)</span> <span class="op">==</span> <span class="fl">3</span><span class="op">)</span>, <span class="op">]</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># A tibble: 1 × 46</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># A tibble: 2 × 46</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> date patient age gender ward mo PEN OXA FLC AMX </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494; font-style: italic;"><date></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><dbl></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><mo></span> <span style="color: #949494; font-style: italic;"><sir></span> <span style="color: #949494; font-style: italic;"><sir></span> <span style="color: #949494; font-style: italic;"><sir></span> <span style="color: #949494; font-style: italic;"><sir></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">1</span> 2002-06-06 24D393 20 F Clinical B_ESCHR_COLI R NA NA NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">1</span> 2002-08-19 A49852 70 M Clinical B_ESCHR_COLI R NA NA NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">2</span> 2002-08-19 A49852 70 M Clinical B_ESCHR_COLI R NA NA NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># ℹ 36 more variables: AMC <sir>, AMP <sir>, TZP <sir>, CZO <sir>, FEP <sir>,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># CXM <sir>, FOX <sir>, CTX <sir>, CAZ <sir>, CRO <sir>, GEN <sir>,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># TOB <sir>, AMK <sir>, KAN <sir>, TMP <sir>, SXT <sir>, NIT <sir>,</span></span>
|
||||
@ -210,16 +211,16 @@
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># Groups: patient, condition [99]</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> patient date condition new_episode</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><date></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><lgl></span> </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 1</span> 006606 2011-10-30 A TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 2</span> 059414 2006-07-21 C TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 3</span> 066895 2002-02-27 C TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 4</span> 067927 2002-01-16 A TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 5</span> 0DBF93 2015-10-12 A TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 6</span> 0F9638 2014-09-22 C TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 7</span> 179451 2007-09-15 B TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 8</span> 183220 2008-11-14 C TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 9</span> 1D4C00 2011-04-04 B TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">10</span> 218456 2003-09-26 C TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 1</span> 001213 2009-08-03 B TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 2</span> 036063 2010-01-28 C TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 3</span> 04C169 2012-07-18 A TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 4</span> 077922 2009-08-18 B TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 5</span> 080086 2007-10-26 A TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 6</span> 088256 2003-01-25 C TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 7</span> 0D7D34 2011-03-19 A TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 8</span> 0DBF93 2015-10-12 A TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 9</span> 122506 2007-08-11 A TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">10</span> 144549 2009-09-01 C TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># ℹ 90 more rows</span></span>
|
||||
<span class="r-in"><span></span></span>
|
||||
<span class="r-in"><span><span class="kw">if</span> <span class="op">(</span><span class="kw"><a href="https://rdrr.io/r/base/library.html" class="external-link">require</a></span><span class="op">(</span><span class="st"><a href="https://dplyr.tidyverse.org" class="external-link">"dplyr"</a></span><span class="op">)</span><span class="op">)</span> <span class="op">{</span></span></span>
|
||||
@ -233,19 +234,19 @@
|
||||
<span class="r-in"><span> <span class="fu"><a href="https://dplyr.tidyverse.org/reference/arrange.html" class="external-link">arrange</a></span><span class="op">(</span><span class="va">patient</span>, <span class="va">ward</span>, <span class="va">date</span><span class="op">)</span></span></span>
|
||||
<span class="r-in"><span><span class="op">}</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># A tibble: 100 × 5</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># Groups: ward, patient [94]</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># Groups: ward, patient [96]</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> ward date patient new_index new_logical</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><date></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><int></span> <span style="color: #949494; font-style: italic;"><lgl></span> </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 1</span> Clinical 2011-10-30 006606 1 TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 2</span> Clinical 2006-07-21 059414 1 TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 3</span> Clinical 2002-02-27 066895 1 TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 4</span> ICU 2002-01-16 067927 1 TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 5</span> Clinical 2015-10-12 0DBF93 1 TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 6</span> Clinical 2014-09-22 0F9638 1 TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 7</span> ICU 2007-09-15 179451 1 TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 8</span> Clinical 2008-11-14 183220 1 TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 9</span> Clinical 2011-04-04 1D4C00 1 TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">10</span> Clinical 2003-09-26 218456 1 TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 1</span> Clinical 2009-08-03 001213 1 TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 2</span> Clinical 2010-01-28 036063 1 TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 3</span> Clinical 2012-07-18 04C169 1 TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 4</span> Clinical 2009-08-18 077922 1 TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 5</span> Clinical 2007-10-26 080086 1 TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 6</span> ICU 2003-01-25 088256 1 TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 7</span> ICU 2011-03-19 0D7D34 1 TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 8</span> Clinical 2015-10-12 0DBF93 1 TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 9</span> Clinical 2007-08-11 122506 1 TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">10</span> Clinical 2009-09-01 144549 1 TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># ℹ 90 more rows</span></span>
|
||||
<span class="r-in"><span></span></span>
|
||||
<span class="r-in"><span><span class="kw">if</span> <span class="op">(</span><span class="kw"><a href="https://rdrr.io/r/base/library.html" class="external-link">require</a></span><span class="op">(</span><span class="st"><a href="https://dplyr.tidyverse.org" class="external-link">"dplyr"</a></span><span class="op">)</span><span class="op">)</span> <span class="op">{</span></span></span>
|
||||
@ -261,9 +262,9 @@
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># A tibble: 3 × 5</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> ward n_patients n_episodes_365 n_episodes_60 n_episodes_30</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><int></span> <span style="color: #949494; font-style: italic;"><int></span> <span style="color: #949494; font-style: italic;"><int></span> <span style="color: #949494; font-style: italic;"><int></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">1</span> Clinical 68 13 40 51</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">2</span> ICU 22 9 17 19</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">3</span> Outpatient 4 3 4 4</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">1</span> Clinical 66 14 41 50</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">2</span> ICU 24 10 17 19</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">3</span> Outpatient 6 4 6 6</span>
|
||||
<span class="r-in"><span></span></span>
|
||||
<span class="r-in"><span><span class="co"># grouping on patients and microorganisms leads to the same</span></span></span>
|
||||
<span class="r-in"><span><span class="co"># results as first_isolate() when using 'episode-based':</span></span></span>
|
||||
@ -292,19 +293,19 @@
|
||||
<span class="r-in"><span> <span class="fu"><a href="https://dplyr.tidyverse.org/reference/select.html" class="external-link">select</a></span><span class="op">(</span><span class="fu"><a href="https://dplyr.tidyverse.org/reference/group_data.html" class="external-link">group_vars</a></span><span class="op">(</span><span class="va">.</span><span class="op">)</span>, <span class="va">flag_episode</span><span class="op">)</span></span></span>
|
||||
<span class="r-in"><span><span class="op">}</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># A tibble: 100 × 4</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># Groups: patient, mo, ward [98]</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># Groups: patient, mo, ward [97]</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> patient mo ward flag_episode</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><mo></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><lgl></span> </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 1</span> F76081 B_ESCHR_COLI ICU TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 2</span> F74547 B_STPHY_EPDR Clinical TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 3</span> 988763 B_STPHY_AURS Clinical TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 4</span> 8DB5B8 B_STPHY_CONS Clinical TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 5</span> FB50D6 B_STRPT_MITS ICU TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 6</span> 310665 B_STPHY_CPTS Outpatient TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 7</span> 3D2C93 B_STPHY_EPDR ICU TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 8</span> D80438 B_CRYNB_STRT Clinical TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 9</span> 24D393 B_ESCHR_COLI Clinical TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">10</span> 066895 B_KLBSL_PNMN Clinical TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 1</span> D80753 B_STPHY_CONS Outpatient TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 2</span> 04C169 B_STPHY_CONS Clinical TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 3</span> D63414 B_PROTS_MRBL Clinical TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 4</span> EE2510 B_ESCHR_COLI ICU TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 5</span> F08866 B_ESCHR_COLI Clinical TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 6</span> A59636 B_STPHY_AURS Clinical TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 7</span> 080086 B_STRPT_GRPB Clinical TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 8</span> 3CF3C4 B_STPHY_CONS Clinical TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 9</span> C80762 B_ESCHR_COLI ICU TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">10</span> 534816 F_CANDD_ALBC Clinical TRUE </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># ℹ 90 more rows</span></span>
|
||||
<span class="r-in"><span><span class="co"># }</span></span></span>
|
||||
</code></pre></div>
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -54,30 +54,13 @@
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">ggplot_pca</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> choices <span class="op">=</span> <span class="fl">1</span><span class="op">:</span><span class="fl">2</span>,</span>
|
||||
<span> scale <span class="op">=</span> <span class="fl">1</span>,</span>
|
||||
<span> pc.biplot <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> labels <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> labels_textsize <span class="op">=</span> <span class="fl">3</span>,</span>
|
||||
<span> labels_text_placement <span class="op">=</span> <span class="fl">1.5</span>,</span>
|
||||
<span> groups <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> ellipse <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> ellipse_prob <span class="op">=</span> <span class="fl">0.68</span>,</span>
|
||||
<span> ellipse_size <span class="op">=</span> <span class="fl">0.5</span>,</span>
|
||||
<span> ellipse_alpha <span class="op">=</span> <span class="fl">0.5</span>,</span>
|
||||
<span> points_size <span class="op">=</span> <span class="fl">2</span>,</span>
|
||||
<span> points_alpha <span class="op">=</span> <span class="fl">0.25</span>,</span>
|
||||
<span> arrows <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> arrows_colour <span class="op">=</span> <span class="st">"darkblue"</span>,</span>
|
||||
<span> arrows_size <span class="op">=</span> <span class="fl">0.5</span>,</span>
|
||||
<span> arrows_textsize <span class="op">=</span> <span class="fl">3</span>,</span>
|
||||
<span> arrows_textangled <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> arrows_alpha <span class="op">=</span> <span class="fl">0.75</span>,</span>
|
||||
<span> base_textsize <span class="op">=</span> <span class="fl">10</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span></code></pre></div>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">ggplot_pca</span><span class="op">(</span><span class="va">x</span>, choices <span class="op">=</span> <span class="fl">1</span><span class="op">:</span><span class="fl">2</span>, scale <span class="op">=</span> <span class="fl">1</span>, pc.biplot <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> labels <span class="op">=</span> <span class="cn">NULL</span>, labels_textsize <span class="op">=</span> <span class="fl">3</span>, labels_text_placement <span class="op">=</span> <span class="fl">1.5</span>,</span>
|
||||
<span> groups <span class="op">=</span> <span class="cn">NULL</span>, ellipse <span class="op">=</span> <span class="cn">TRUE</span>, ellipse_prob <span class="op">=</span> <span class="fl">0.68</span>,</span>
|
||||
<span> ellipse_size <span class="op">=</span> <span class="fl">0.5</span>, ellipse_alpha <span class="op">=</span> <span class="fl">0.5</span>, points_size <span class="op">=</span> <span class="fl">2</span>,</span>
|
||||
<span> points_alpha <span class="op">=</span> <span class="fl">0.25</span>, arrows <span class="op">=</span> <span class="cn">TRUE</span>, arrows_colour <span class="op">=</span> <span class="st">"darkblue"</span>,</span>
|
||||
<span> arrows_size <span class="op">=</span> <span class="fl">0.5</span>, arrows_textsize <span class="op">=</span> <span class="fl">3</span>, arrows_textangled <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> arrows_alpha <span class="op">=</span> <span class="fl">0.75</span>, base_textsize <span class="op">=</span> <span class="fl">10</span>, <span class="va">...</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -54,42 +54,18 @@
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">ggplot_sir</span><span class="op">(</span></span>
|
||||
<span> <span class="va">data</span>,</span>
|
||||
<span> position <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> x <span class="op">=</span> <span class="st">"antibiotic"</span>,</span>
|
||||
<span> fill <span class="op">=</span> <span class="st">"interpretation"</span>,</span>
|
||||
<span> facet <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> breaks <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/seq.html" class="external-link">seq</a></span><span class="op">(</span><span class="fl">0</span>, <span class="fl">1</span>, <span class="fl">0.1</span><span class="op">)</span>,</span>
|
||||
<span> limits <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> translate_ab <span class="op">=</span> <span class="st">"name"</span>,</span>
|
||||
<span> combine_SI <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> minimum <span class="op">=</span> <span class="fl">30</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> nrow <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> colours <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span>S <span class="op">=</span> <span class="st">"#3CAEA3"</span>, SI <span class="op">=</span> <span class="st">"#3CAEA3"</span>, I <span class="op">=</span> <span class="st">"#F6D55C"</span>, IR <span class="op">=</span> <span class="st">"#ED553B"</span>, R <span class="op">=</span></span>
|
||||
<span> <span class="st">"#ED553B"</span><span class="op">)</span>,</span>
|
||||
<span> datalabels <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> datalabels.size <span class="op">=</span> <span class="fl">2.5</span>,</span>
|
||||
<span> datalabels.colour <span class="op">=</span> <span class="st">"grey15"</span>,</span>
|
||||
<span> title <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> subtitle <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> caption <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> x.title <span class="op">=</span> <span class="st">"Antimicrobial"</span>,</span>
|
||||
<span> y.title <span class="op">=</span> <span class="st">"Proportion"</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">ggplot_sir</span><span class="op">(</span><span class="va">data</span>, position <span class="op">=</span> <span class="cn">NULL</span>, x <span class="op">=</span> <span class="st">"antibiotic"</span>,</span>
|
||||
<span> fill <span class="op">=</span> <span class="st">"interpretation"</span>, facet <span class="op">=</span> <span class="cn">NULL</span>, breaks <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/seq.html" class="external-link">seq</a></span><span class="op">(</span><span class="fl">0</span>, <span class="fl">1</span>, <span class="fl">0.1</span><span class="op">)</span>,</span>
|
||||
<span> limits <span class="op">=</span> <span class="cn">NULL</span>, translate_ab <span class="op">=</span> <span class="st">"name"</span>, combine_SI <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> minimum <span class="op">=</span> <span class="fl">30</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>, nrow <span class="op">=</span> <span class="cn">NULL</span>, colours <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span>S</span>
|
||||
<span> <span class="op">=</span> <span class="st">"#3CAEA3"</span>, SI <span class="op">=</span> <span class="st">"#3CAEA3"</span>, I <span class="op">=</span> <span class="st">"#F6D55C"</span>, IR <span class="op">=</span> <span class="st">"#ED553B"</span>, R <span class="op">=</span> <span class="st">"#ED553B"</span><span class="op">)</span>,</span>
|
||||
<span> datalabels <span class="op">=</span> <span class="cn">TRUE</span>, datalabels.size <span class="op">=</span> <span class="fl">2.5</span>, datalabels.colour <span class="op">=</span> <span class="st">"grey15"</span>,</span>
|
||||
<span> title <span class="op">=</span> <span class="cn">NULL</span>, subtitle <span class="op">=</span> <span class="cn">NULL</span>, caption <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> x.title <span class="op">=</span> <span class="st">"Antimicrobial"</span>, y.title <span class="op">=</span> <span class="st">"Proportion"</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">geom_sir</span><span class="op">(</span></span>
|
||||
<span> position <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> x <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"antibiotic"</span>, <span class="st">"interpretation"</span><span class="op">)</span>,</span>
|
||||
<span> fill <span class="op">=</span> <span class="st">"interpretation"</span>,</span>
|
||||
<span> translate_ab <span class="op">=</span> <span class="st">"name"</span>,</span>
|
||||
<span> minimum <span class="op">=</span> <span class="fl">30</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> combine_SI <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span></code></pre></div>
|
||||
<span><span class="fu">geom_sir</span><span class="op">(</span>position <span class="op">=</span> <span class="cn">NULL</span>, x <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"antibiotic"</span>, <span class="st">"interpretation"</span><span class="op">)</span>,</span>
|
||||
<span> fill <span class="op">=</span> <span class="st">"interpretation"</span>, translate_ab <span class="op">=</span> <span class="st">"name"</span>, minimum <span class="op">=</span> <span class="fl">30</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>, combine_SI <span class="op">=</span> <span class="cn">TRUE</span>, <span class="va">...</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -54,12 +54,8 @@
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">guess_ab_col</span><span class="op">(</span></span>
|
||||
<span> x <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> search_string <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> verbose <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span></span>
|
||||
<span><span class="op">)</span></span></code></pre></div>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">guess_ab_col</span><span class="op">(</span>x <span class="op">=</span> <span class="cn">NULL</span>, search_string <span class="op">=</span> <span class="cn">NULL</span>, verbose <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -183,7 +183,7 @@
|
||||
</dl></div><div class="section level2">
|
||||
<h2 id="analysing-data">Analysing data<a class="anchor" aria-label="anchor" href="#analysing-data"></a></h2>
|
||||
|
||||
<div class="section-desc"><p>Use these function for the analysis part. You can use <code><a href="../reference/proportion.html">susceptibility()</a></code> or <code><a href="../reference/proportion.html">resistance()</a></code> on any antibiotic column. With <code><a href="../reference/antibiogram.html">antibiogram()</a></code>, you can generate a traditional, combined, syndromic, or weighted-incidence syndromic combination antibiogram(WISCA). This function also comes with support for R Markdown and Quarto. Be sure to first select the isolates that are appropiate for analysis, by using <code><a href="../reference/first_isolate.html">first_isolate()</a></code> or <code><a href="../reference/get_episode.html">is_new_episode()</a></code>. You can also filter your data on certain resistance in certain antibiotic classes (<code><a href="../reference/antimicrobial_class_selectors.html">carbapenems()</a></code>, <code><a href="../reference/antimicrobial_class_selectors.html">aminoglycosides()</a></code>), or determine multi-drug resistant microorganisms (MDRO, <code><a href="../reference/mdro.html">mdro()</a></code>).</p></div>
|
||||
<div class="section-desc"><p>Use these function for the analysis part. You can use <code><a href="../reference/proportion.html">susceptibility()</a></code> or <code><a href="../reference/proportion.html">resistance()</a></code> on any antibiotic column. With <code><a href="../reference/antibiogram.html">antibiogram()</a></code>, you can generate a traditional, combined, syndromic, or weighted-incidence syndromic combination antibiogram (WISCA). This function also comes with support for R Markdown and Quarto. Be sure to first select the isolates that are appropiate for analysis, by using <code><a href="../reference/first_isolate.html">first_isolate()</a></code> or <code><a href="../reference/get_episode.html">is_new_episode()</a></code>. You can also filter your data on certain resistance in certain antibiotic classes (<code><a href="../reference/antimicrobial_class_selectors.html">carbapenems()</a></code>, <code><a href="../reference/antimicrobial_class_selectors.html">aminoglycosides()</a></code>), or determine multi-drug resistant microorganisms (MDRO, <code><a href="../reference/mdro.html">mdro()</a></code>).</p></div>
|
||||
|
||||
|
||||
</div><div class="section level2">
|
||||
@ -193,7 +193,7 @@
|
||||
|
||||
<dl><dt>
|
||||
|
||||
<code><a href="antibiogram.html">antibiogram()</a></code> <code><a href="antibiogram.html">plot(<i><antibiogram></i>)</a></code> <code><a href="antibiogram.html">autoplot(<i><antibiogram></i>)</a></code> <code><a href="antibiogram.html">knit_print(<i><antibiogram></i>)</a></code>
|
||||
<code><a href="antibiogram.html">antibiogram()</a></code> <code><a href="antibiogram.html">wisca()</a></code> <code><a href="antibiogram.html">plot(<i><antibiogram></i>)</a></code> <code><a href="antibiogram.html">autoplot(<i><antibiogram></i>)</a></code> <code><a href="antibiogram.html">knit_print(<i><antibiogram></i>)</a></code>
|
||||
|
||||
</dt>
|
||||
<dd>Generate Traditional, Combination, Syndromic, or WISCA Antibiograms</dd>
|
||||
@ -247,6 +247,12 @@
|
||||
<dd>Antimicrobial Selectors</dd>
|
||||
</dl><dl><dt>
|
||||
|
||||
<code><a href="top_n_microorganisms.html">top_n_microorganisms()</a></code>
|
||||
|
||||
</dt>
|
||||
<dd>Filter Top <em>n</em> Microorganisms</dd>
|
||||
</dl><dl><dt>
|
||||
|
||||
<code><a href="mean_amr_distance.html">mean_amr_distance()</a></code> <code><a href="mean_amr_distance.html">amr_distance_from_row()</a></code>
|
||||
|
||||
</dt>
|
||||
@ -521,6 +527,23 @@
|
||||
|
||||
</dt>
|
||||
<dd>Skewness of the Sample</dd>
|
||||
</dl></div><div class="section level2">
|
||||
<h2 id="other-deprecated-functions">Other: deprecated functions<a class="anchor" aria-label="anchor" href="#other-deprecated-functions"></a></h2>
|
||||
|
||||
<div class="section-desc"><p>These functions are deprecated, meaning that they will still work but show a warning that they will be removed in a future version.</p></div>
|
||||
|
||||
|
||||
</div><div class="section level2">
|
||||
|
||||
|
||||
|
||||
|
||||
<dl><dt>
|
||||
|
||||
<code><a href="AMR-deprecated.html">ab_class()</a></code> <code><a href="AMR-deprecated.html">ab_selector()</a></code>
|
||||
|
||||
</dt>
|
||||
<dd>Deprecated Functions</dd>
|
||||
</dl></div>
|
||||
</main><aside class="col-md-3"><nav id="toc" aria-label="Table of contents"><h2>On this page</h2>
|
||||
</nav></aside></div>
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -54,31 +54,19 @@
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">key_antimicrobials</span><span class="op">(</span></span>
|
||||
<span> x <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> col_mo <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> universal <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"ampicillin"</span>, <span class="st">"amoxicillin/clavulanic acid"</span>, <span class="st">"cefuroxime"</span>,</span>
|
||||
<span> <span class="st">"piperacillin/tazobactam"</span>, <span class="st">"ciprofloxacin"</span>, <span class="st">"trimethoprim/sulfamethoxazole"</span><span class="op">)</span>,</span>
|
||||
<span> gram_negative <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"gentamicin"</span>, <span class="st">"tobramycin"</span>, <span class="st">"colistin"</span>, <span class="st">"cefotaxime"</span>, <span class="st">"ceftazidime"</span>,</span>
|
||||
<span> <span class="st">"meropenem"</span><span class="op">)</span>,</span>
|
||||
<span> gram_positive <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"vancomycin"</span>, <span class="st">"teicoplanin"</span>, <span class="st">"tetracycline"</span>, <span class="st">"erythromycin"</span>,</span>
|
||||
<span> <span class="st">"oxacillin"</span>, <span class="st">"rifampin"</span><span class="op">)</span>,</span>
|
||||
<span> antifungal <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"anidulafungin"</span>, <span class="st">"caspofungin"</span>, <span class="st">"fluconazole"</span>, <span class="st">"miconazole"</span>, <span class="st">"nystatin"</span>,</span>
|
||||
<span> <span class="st">"voriconazole"</span><span class="op">)</span>,</span>
|
||||
<span> only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">key_antimicrobials</span><span class="op">(</span>x <span class="op">=</span> <span class="cn">NULL</span>, col_mo <span class="op">=</span> <span class="cn">NULL</span>, universal <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"ampicillin"</span>,</span>
|
||||
<span> <span class="st">"amoxicillin/clavulanic acid"</span>, <span class="st">"cefuroxime"</span>, <span class="st">"piperacillin/tazobactam"</span>,</span>
|
||||
<span> <span class="st">"ciprofloxacin"</span>, <span class="st">"trimethoprim/sulfamethoxazole"</span><span class="op">)</span>,</span>
|
||||
<span> gram_negative <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"gentamicin"</span>, <span class="st">"tobramycin"</span>, <span class="st">"colistin"</span>, <span class="st">"cefotaxime"</span>,</span>
|
||||
<span> <span class="st">"ceftazidime"</span>, <span class="st">"meropenem"</span><span class="op">)</span>, gram_positive <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"vancomycin"</span>, <span class="st">"teicoplanin"</span>,</span>
|
||||
<span> <span class="st">"tetracycline"</span>, <span class="st">"erythromycin"</span>, <span class="st">"oxacillin"</span>, <span class="st">"rifampin"</span><span class="op">)</span>,</span>
|
||||
<span> antifungal <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"anidulafungin"</span>, <span class="st">"caspofungin"</span>, <span class="st">"fluconazole"</span>, <span class="st">"miconazole"</span>,</span>
|
||||
<span> <span class="st">"nystatin"</span>, <span class="st">"voriconazole"</span><span class="op">)</span>, only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">all_antimicrobials</span><span class="op">(</span>x <span class="op">=</span> <span class="cn">NULL</span>, only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">antimicrobials_equal</span><span class="op">(</span></span>
|
||||
<span> <span class="va">y</span>,</span>
|
||||
<span> <span class="va">z</span>,</span>
|
||||
<span> type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"points"</span>, <span class="st">"keyantimicrobials"</span><span class="op">)</span>,</span>
|
||||
<span> ignore_I <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> points_threshold <span class="op">=</span> <span class="fl">2</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span></code></pre></div>
|
||||
<span><span class="fu">antimicrobials_equal</span><span class="op">(</span><span class="va">y</span>, <span class="va">z</span>, type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"points"</span>, <span class="st">"keyantimicrobials"</span><span class="op">)</span>,</span>
|
||||
<span> ignore_I <span class="op">=</span> <span class="cn">TRUE</span>, points_threshold <span class="op">=</span> <span class="fl">2</span>, <span class="va">...</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -90,9 +90,9 @@
|
||||
<div class="section level2">
|
||||
<h2 id="ref-examples">Examples<a class="anchor" aria-label="anchor" href="#ref-examples"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span class="r-in"><span><span class="fu">kurtosis</span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/stats/Normal.html" class="external-link">rnorm</a></span><span class="op">(</span><span class="fl">10000</span><span class="op">)</span><span class="op">)</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 3.03269</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 2.883046</span>
|
||||
<span class="r-in"><span><span class="fu">kurtosis</span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/stats/Normal.html" class="external-link">rnorm</a></span><span class="op">(</span><span class="fl">10000</span><span class="op">)</span>, excess <span class="op">=</span> <span class="cn">TRUE</span><span class="op">)</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 0.07248166</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] -0.05230069</span>
|
||||
</code></pre></div>
|
||||
</div>
|
||||
</main><aside class="col-md-3"><nav id="toc" aria-label="Table of contents"><h2>On this page</h2>
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -54,23 +54,10 @@
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">mdro</span><span class="op">(</span></span>
|
||||
<span> x <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> guideline <span class="op">=</span> <span class="st">"CMI2012"</span>,</span>
|
||||
<span> col_mo <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> esbl <span class="op">=</span> <span class="cn">NA</span>,</span>
|
||||
<span> carbapenemase <span class="op">=</span> <span class="cn">NA</span>,</span>
|
||||
<span> mecA <span class="op">=</span> <span class="cn">NA</span>,</span>
|
||||
<span> mecC <span class="op">=</span> <span class="cn">NA</span>,</span>
|
||||
<span> vanA <span class="op">=</span> <span class="cn">NA</span>,</span>
|
||||
<span> vanB <span class="op">=</span> <span class="cn">NA</span>,</span>
|
||||
<span> info <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/interactive.html" class="external-link">interactive</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> pct_required_classes <span class="op">=</span> <span class="fl">0.5</span>,</span>
|
||||
<span> combine_SI <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> verbose <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">mdro</span><span class="op">(</span>x <span class="op">=</span> <span class="cn">NULL</span>, guideline <span class="op">=</span> <span class="st">"CMI2012"</span>, col_mo <span class="op">=</span> <span class="cn">NULL</span>, esbl <span class="op">=</span> <span class="cn">NA</span>,</span>
|
||||
<span> carbapenemase <span class="op">=</span> <span class="cn">NA</span>, mecA <span class="op">=</span> <span class="cn">NA</span>, mecC <span class="op">=</span> <span class="cn">NA</span>, vanA <span class="op">=</span> <span class="cn">NA</span>, vanB <span class="op">=</span> <span class="cn">NA</span>,</span>
|
||||
<span> info <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/interactive.html" class="external-link">interactive</a></span><span class="op">(</span><span class="op">)</span>, pct_required_classes <span class="op">=</span> <span class="fl">0.5</span>, combine_SI <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> verbose <span class="op">=</span> <span class="cn">FALSE</span>, only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">custom_mdro_guideline</span><span class="op">(</span><span class="va">...</span>, as_factor <span class="op">=</span> <span class="cn">TRUE</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
@ -82,12 +69,8 @@
|
||||
<span></span>
|
||||
<span><span class="fu">mdr_cmi2012</span><span class="op">(</span>x <span class="op">=</span> <span class="cn">NULL</span>, only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>, verbose <span class="op">=</span> <span class="cn">FALSE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">eucast_exceptional_phenotypes</span><span class="op">(</span></span>
|
||||
<span> x <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> verbose <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span></code></pre></div>
|
||||
<span><span class="fu">eucast_exceptional_phenotypes</span><span class="op">(</span>x <span class="op">=</span> <span class="cn">NULL</span>, only_sir_columns <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> verbose <span class="op">=</span> <span class="cn">FALSE</span>, <span class="va">...</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -109,30 +109,31 @@
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span class="r-in"><span><span class="va">sir</span> <span class="op"><-</span> <span class="fu"><a href="random.html">random_sir</a></span><span class="op">(</span><span class="fl">10</span><span class="op">)</span></span></span>
|
||||
<span class="r-in"><span><span class="va">sir</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> Class 'sir'</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] I I S I I S S S R R</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] S I S I I I R R I R</span>
|
||||
<span class="r-in"><span><span class="fu">mean_amr_distance</span><span class="op">(</span><span class="va">sir</span><span class="op">)</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] -0.4743416 -0.4743416 -0.4743416 -0.4743416 -0.4743416 -0.4743416</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [7] -0.4743416 -0.4743416 1.8973666 1.8973666</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] -0.621059 -0.621059 -0.621059 -0.621059 -0.621059 -0.621059 1.449138</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [8] 1.449138 -0.621059 1.449138</span>
|
||||
<span class="r-in"><span></span></span>
|
||||
<span class="r-in"><span><span class="va">mic</span> <span class="op"><-</span> <span class="fu"><a href="random.html">random_mic</a></span><span class="op">(</span><span class="fl">10</span><span class="op">)</span></span></span>
|
||||
<span class="r-in"><span><span class="va">mic</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> Class 'mic'</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 0.001 256 0.25 0.5 32 0.002 8 0.125 0.002 0.125</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 64 16 8 64 256 0.5 128 128 2 </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [10] <=0.002</span>
|
||||
<span class="r-in"><span><span class="fu">mean_amr_distance</span><span class="op">(</span><span class="va">mic</span><span class="op">)</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] -1.2863106 1.6478483 0.0146555 0.1779748 1.1578905 -1.1229914</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [7] 0.8312519 -0.1486638 -1.1229914 -0.1486638</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 0.54295251 0.15465479 -0.03949407 0.54295251 0.93125022 -0.81608950</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [7] 0.73710137 0.73710137 -0.42779178 -2.36263742</span>
|
||||
<span class="r-in"><span><span class="co"># equal to the Z-score of their log2:</span></span></span>
|
||||
<span class="r-in"><span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/Log.html" class="external-link">log2</a></span><span class="op">(</span><span class="va">mic</span><span class="op">)</span> <span class="op">-</span> <span class="fu"><a href="https://rdrr.io/r/base/mean.html" class="external-link">mean</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/Log.html" class="external-link">log2</a></span><span class="op">(</span><span class="va">mic</span><span class="op">)</span><span class="op">)</span><span class="op">)</span> <span class="op">/</span> <span class="fu"><a href="https://rdrr.io/r/stats/sd.html" class="external-link">sd</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/Log.html" class="external-link">log2</a></span><span class="op">(</span><span class="va">mic</span><span class="op">)</span><span class="op">)</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] -1.2863106 1.6478483 0.0146555 0.1779748 1.1578905 -1.1229914</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [7] 0.8312519 -0.1486638 -1.1229914 -0.1486638</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 0.54295251 0.15465479 -0.03949407 0.54295251 0.93125022 -0.81608950</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [7] 0.73710137 0.73710137 -0.42779178 -2.36263742</span>
|
||||
<span class="r-in"><span></span></span>
|
||||
<span class="r-in"><span><span class="va">disk</span> <span class="op"><-</span> <span class="fu"><a href="random.html">random_disk</a></span><span class="op">(</span><span class="fl">10</span><span class="op">)</span></span></span>
|
||||
<span class="r-in"><span><span class="va">disk</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> Class 'disk'</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 41 29 40 12 16 7 45 46 22 9</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 28 27 30 48 9 45 12 26 42 25</span>
|
||||
<span class="r-in"><span><span class="fu">mean_amr_distance</span><span class="op">(</span><span class="va">disk</span><span class="op">)</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 0.9256143 0.1488750 0.8608860 -0.9515056 -0.6925925 -1.2751469</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [7] 1.1845274 1.2492556 -0.3042229 -1.1456904</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] -0.09277857 -0.17009405 0.06185238 1.45353101 -1.56177268 1.22158457</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [7] -1.32982624 -0.24740953 0.98963813 -0.32472501</span>
|
||||
<span class="r-in"><span></span></span>
|
||||
<span class="r-in"><span><span class="va">y</span> <span class="op"><-</span> <span class="fu"><a href="https://rdrr.io/r/base/data.frame.html" class="external-link">data.frame</a></span><span class="op">(</span></span></span>
|
||||
<span class="r-in"><span> id <span class="op">=</span> <span class="va">LETTERS</span><span class="op">[</span><span class="fl">1</span><span class="op">:</span><span class="fl">10</span><span class="op">]</span>,</span></span>
|
||||
@ -142,22 +143,22 @@
|
||||
<span class="r-in"><span> tobr <span class="op">=</span> <span class="fu"><a href="random.html">random_mic</a></span><span class="op">(</span><span class="fl">10</span>, ab <span class="op">=</span> <span class="st">"tobr"</span>, mo <span class="op">=</span> <span class="st">"Escherichia coli"</span><span class="op">)</span></span></span>
|
||||
<span class="r-in"><span><span class="op">)</span></span></span>
|
||||
<span class="r-in"><span><span class="va">y</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> id amox cipr gent tobr</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 1 A R 29 <=1 4</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 2 B I 30 4 <=2</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 3 C I 26 16 16</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 4 D I 18 2 8</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 5 E S 29 4 4</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 6 F R 31 16 <=2</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 7 G S 31 <=1 16</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 8 H R 21 16 16</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 9 I I 31 16 16</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 10 J S 21 <=1 4</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> id amox cipr gent tobr</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 1 A I 28 2 0.5</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 2 B R 29 1 4</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 3 C S 23 4 4</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 4 D I 27 4 2</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 5 E S 19 1 4</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 6 F S 22 0.5 <=0.25</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 7 G I 26 1 4</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 8 H R 17 2 0.5</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 9 I S 19 1 1</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 10 J I 30 2 2</span>
|
||||
<span class="r-in"><span><span class="fu">mean_amr_distance</span><span class="op">(</span><span class="va">y</span><span class="op">)</span></span></span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> ℹ Calculating mean AMR distance based on columns "amox", "cipr", "gent",</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> and "tobr"</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 0.04629994 -0.34117383 0.33391321 -0.69063550 -0.19222473 0.50618460</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [7] 0.02978803 0.59753857 0.58783703 -0.87752733</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] -0.06537260 0.82078512 0.42232605 0.47166071 -0.31061057 -1.07205573</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [7] 0.06631747 -0.06476102 -0.64271615 0.37442671</span>
|
||||
<span class="r-in"><span><span class="va">y</span><span class="op">$</span><span class="va">amr_distance</span> <span class="op"><-</span> <span class="fu">mean_amr_distance</span><span class="op">(</span><span class="va">y</span>, <span class="fu"><a href="https://tidyselect.r-lib.org/reference/where.html" class="external-link">where</a></span><span class="op">(</span><span class="va">is.mic</span><span class="op">)</span><span class="op">)</span></span></span>
|
||||
<span class="r-err co"><span class="r-pr">#></span> <span class="error">Error in .subset(x, j):</span> invalid subscript type 'list'</span>
|
||||
<span class="r-in"><span><span class="va">y</span><span class="op">[</span><span class="fu"><a href="https://rdrr.io/r/base/order.html" class="external-link">order</a></span><span class="op">(</span><span class="va">y</span><span class="op">$</span><span class="va">amr_distance</span><span class="op">)</span>, <span class="op">]</span></span></span>
|
||||
@ -173,17 +174,17 @@
|
||||
<span class="r-in"><span><span class="op">}</span></span></span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> ℹ Calculating mean AMR distance based on columns "amox", "cipr", "gent",</span>
|
||||
<span class="r-msg co"><span class="r-pr">#></span> and "tobr"</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> id amox cipr gent tobr amr_distance check_id_C</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 1 C I 26 16 16 0.33391321 0.0000000</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 2 F R 31 16 <=2 0.50618460 0.1722714</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 3 I I 31 16 16 0.58783703 0.2539238</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 4 H R 21 16 16 0.59753857 0.2636254</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 5 A R 29 <=1 4 0.04629994 0.2876133</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 6 G S 31 <=1 16 0.02978803 0.3041252</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 7 E S 29 4 4 -0.19222473 0.5261379</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 8 B I 30 4 <=2 -0.34117383 0.6750870</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 9 D I 18 2 8 -0.69063550 1.0245487</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 10 J S 21 <=1 4 -0.87752733 1.2114405</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> id amox cipr gent tobr amr_distance check_id_C</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 1 C S 23 4 4 0.42232605 0.00000000</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 2 J I 30 2 2 0.37442671 0.04789934</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 3 D I 27 4 2 0.47166071 0.04933466</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 4 G I 26 1 4 0.06631747 0.35600858</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 5 B R 29 1 4 0.82078512 0.39845906</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 6 H R 17 2 0.5 -0.06476102 0.48708707</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 7 A I 28 2 0.5 -0.06537260 0.48769865</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 8 E S 19 1 4 -0.31061057 0.73293662</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 9 I S 19 1 1 -0.64271615 1.06504220</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> 10 F S 22 0.5 <=0.25 -1.07205573 1.49438178</span>
|
||||
<span class="r-in"><span><span class="kw">if</span> <span class="op">(</span><span class="kw"><a href="https://rdrr.io/r/base/library.html" class="external-link">require</a></span><span class="op">(</span><span class="st"><a href="https://dplyr.tidyverse.org" class="external-link">"dplyr"</a></span><span class="op">)</span><span class="op">)</span> <span class="op">{</span></span></span>
|
||||
<span class="r-in"><span> <span class="co"># support for groups</span></span></span>
|
||||
<span class="r-in"><span> <span class="va">example_isolates</span> <span class="op"><a href="https://magrittr.tidyverse.org/reference/pipe.html" class="external-link">%>%</a></span></span></span>
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -9,7 +9,7 @@ This data set is carefully crafted, yet made 100% reproducible from public and a
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -54,262 +54,115 @@
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">mo_name</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">mo_name</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_fullname</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_fullname</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_shortname</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_shortname</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_subspecies</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_subspecies</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_species</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_species</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_genus</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_genus</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_family</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_family</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_order</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_order</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_class</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_class</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_phylum</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_phylum</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_kingdom</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_kingdom</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_domain</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_domain</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_type</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_type</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_status</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_status</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_pathogenicity</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_pathogenicity</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_gramstain</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_gramstain</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_is_gram_negative</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_is_gram_negative</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_is_gram_positive</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_is_gram_positive</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_is_yeast</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_is_yeast</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_is_intrinsic_resistant</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> <span class="va">ab</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_is_intrinsic_resistant</span><span class="op">(</span><span class="va">x</span>, <span class="va">ab</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_oxygen_tolerance</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_oxygen_tolerance</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_is_anaerobic</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_is_anaerobic</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_snomed</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_snomed</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_ref</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_ref</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_authors</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_authors</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_year</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_year</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_lpsn</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_lpsn</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_mycobank</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_mycobank</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_gbif</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_gbif</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_rank</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_rank</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_taxonomy</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_taxonomy</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_synonyms</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_synonyms</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_current</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_group_members</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_group_members</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_info</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_info</span><span class="op">(</span><span class="va">x</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_url</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> open <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">mo_url</span><span class="op">(</span><span class="va">x</span>, open <span class="op">=</span> <span class="cn">FALSE</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">mo_property</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> property <span class="op">=</span> <span class="st">"fullname"</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span></code></pre></div>
|
||||
<span><span class="fu">mo_property</span><span class="op">(</span><span class="va">x</span>, property <span class="op">=</span> <span class="st">"fullname"</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> keep_synonyms <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_keep_synonyms"</span>, <span class="cn">FALSE</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
|
||||
@ -9,7 +9,7 @@ This is the fastest way to have your organisation (or analysis) specific codes p
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -57,10 +57,8 @@ This is the fastest way to have your organisation (or analysis) specific codes p
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">set_mo_source</span><span class="op">(</span></span>
|
||||
<span> <span class="va">path</span>,</span>
|
||||
<span> destination <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_mo_source"</span>, <span class="st">"~/mo_source.rds"</span><span class="op">)</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">set_mo_source</span><span class="op">(</span><span class="va">path</span>, destination <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_mo_source"</span>,</span>
|
||||
<span> <span class="st">"~/mo_source.rds"</span><span class="op">)</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">get_mo_source</span><span class="op">(</span>destination <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_mo_source"</span>, <span class="st">"~/mo_source.rds"</span><span class="op">)</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -54,15 +54,8 @@
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">pca</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> <span class="va">...</span>,</span>
|
||||
<span> retx <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> center <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> scale. <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> tol <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> rank. <span class="op">=</span> <span class="cn">NULL</span></span>
|
||||
<span><span class="op">)</span></span></code></pre></div>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">pca</span><span class="op">(</span><span class="va">x</span>, <span class="va">...</span>, retx <span class="op">=</span> <span class="cn">TRUE</span>, center <span class="op">=</span> <span class="cn">TRUE</span>, scale. <span class="op">=</span> <span class="cn">TRUE</span>, tol <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> rank. <span class="op">=</span> <span class="cn">NULL</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
|
||||
|
Before 
(image error) Size: 26 KiB After 
(image error) Size: 26 KiB 

|
|
Before 
(image error) Size: 37 KiB After 
(image error) Size: 37 KiB 

|
|
Before 
(image error) Size: 54 KiB After 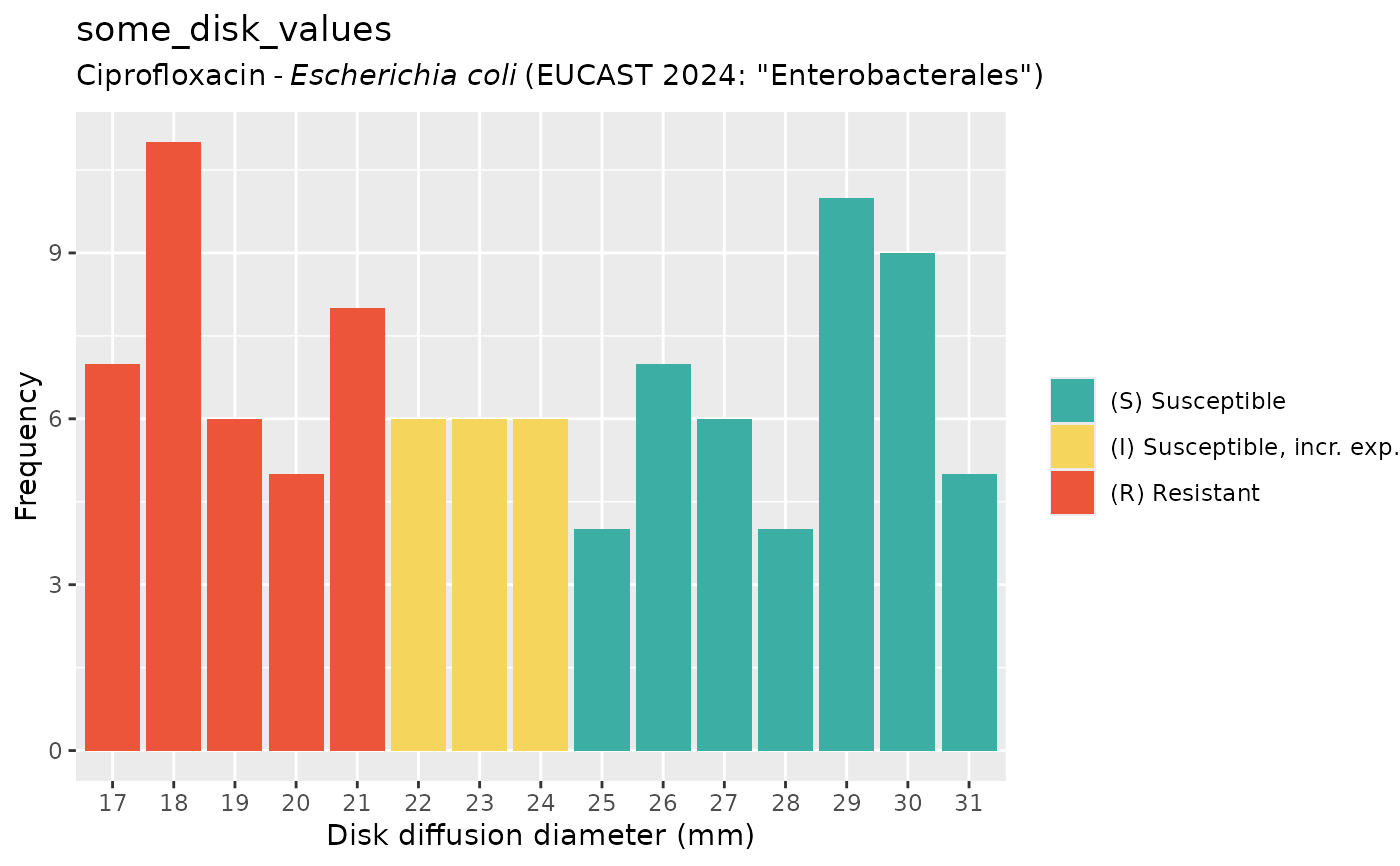
(image error) Size: 54 KiB 

|
|
Before 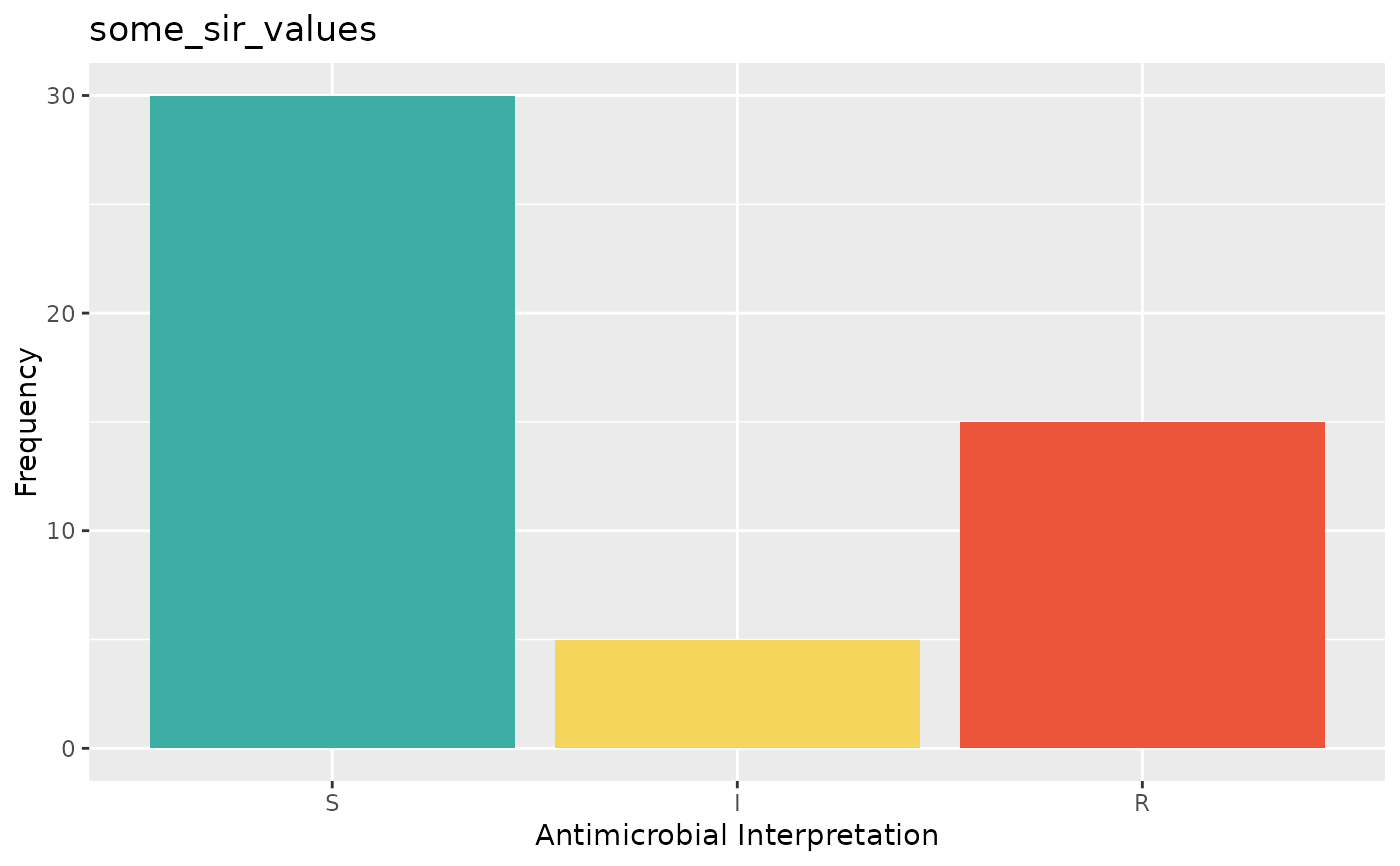
(image error) Size: 25 KiB After 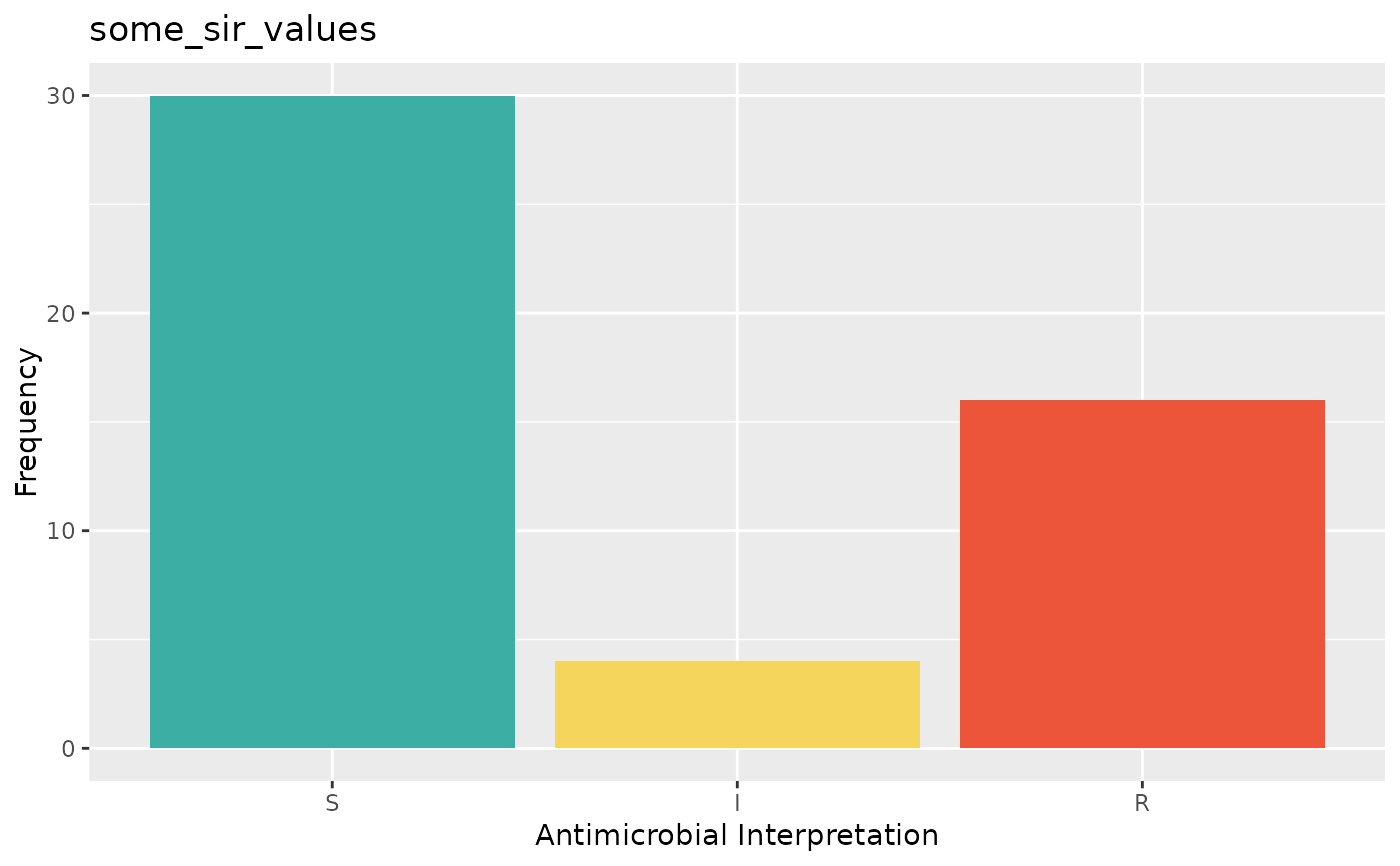
(image error) Size: 25 KiB 

|
|
Before 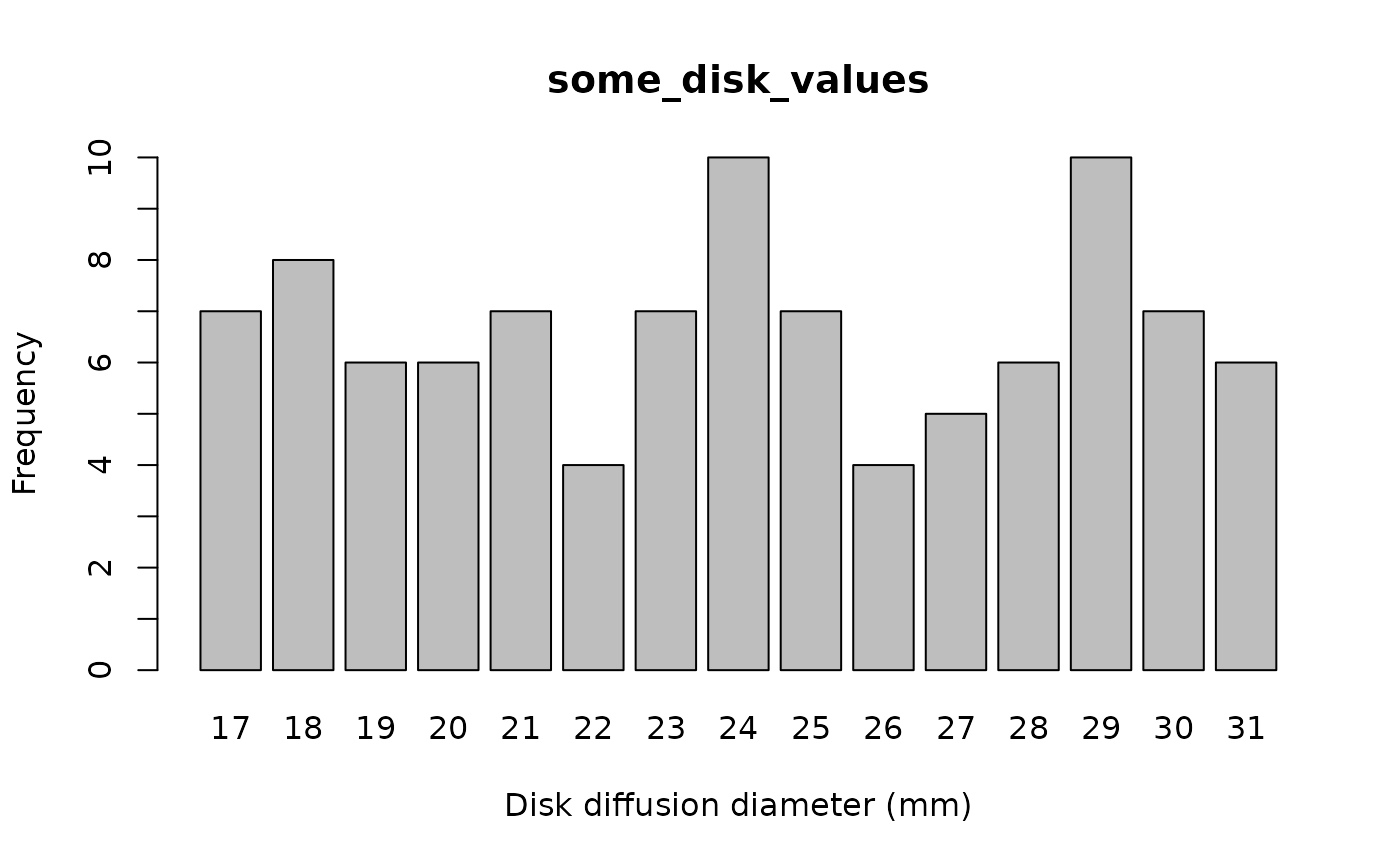
(image error) Size: 26 KiB After 
(image error) Size: 26 KiB 

|
|
Before 
(image error) Size: 28 KiB After 
(image error) Size: 28 KiB 

|
|
Before 
(image error) Size: 39 KiB After 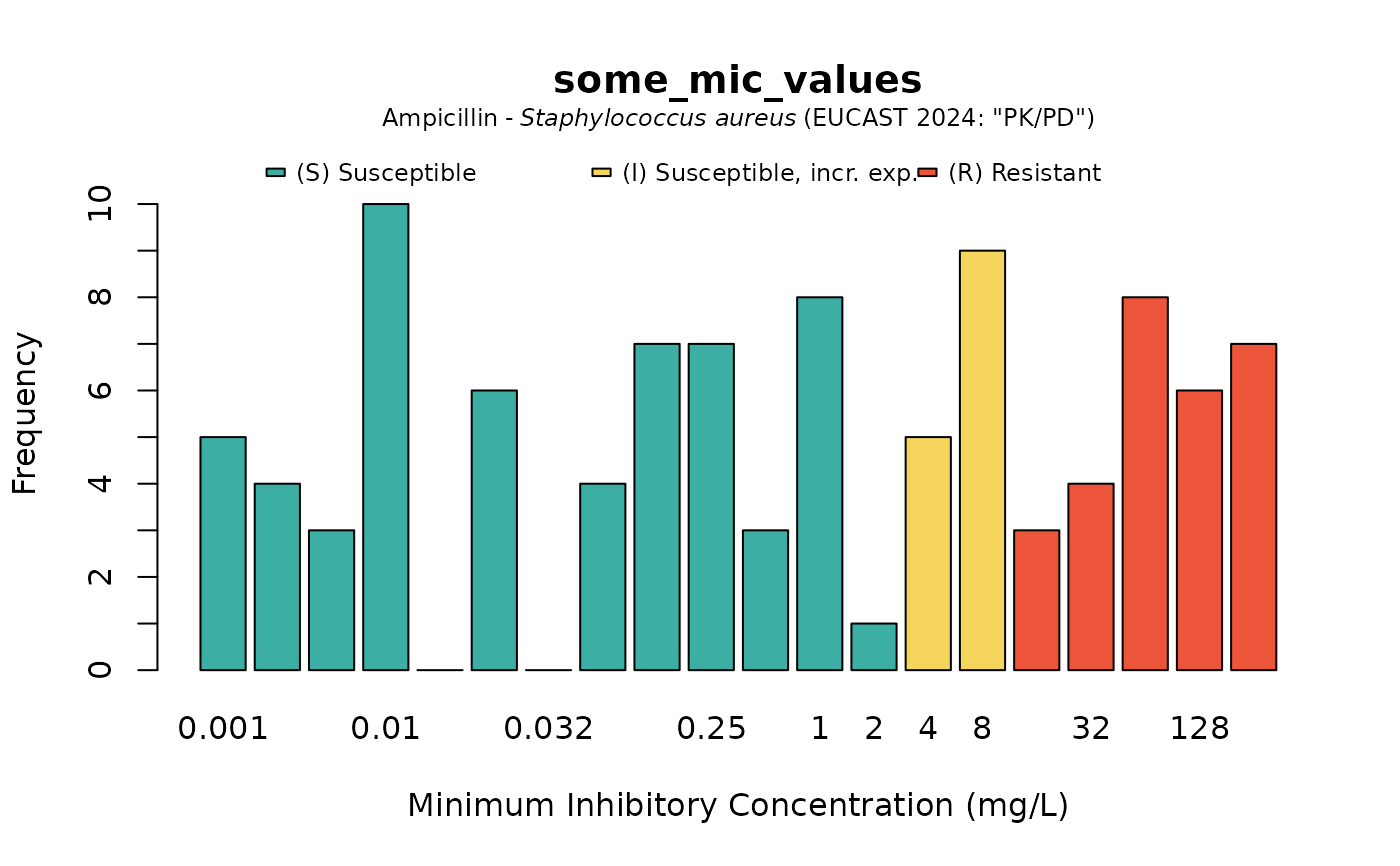
(image error) Size: 39 KiB 

|
|
Before 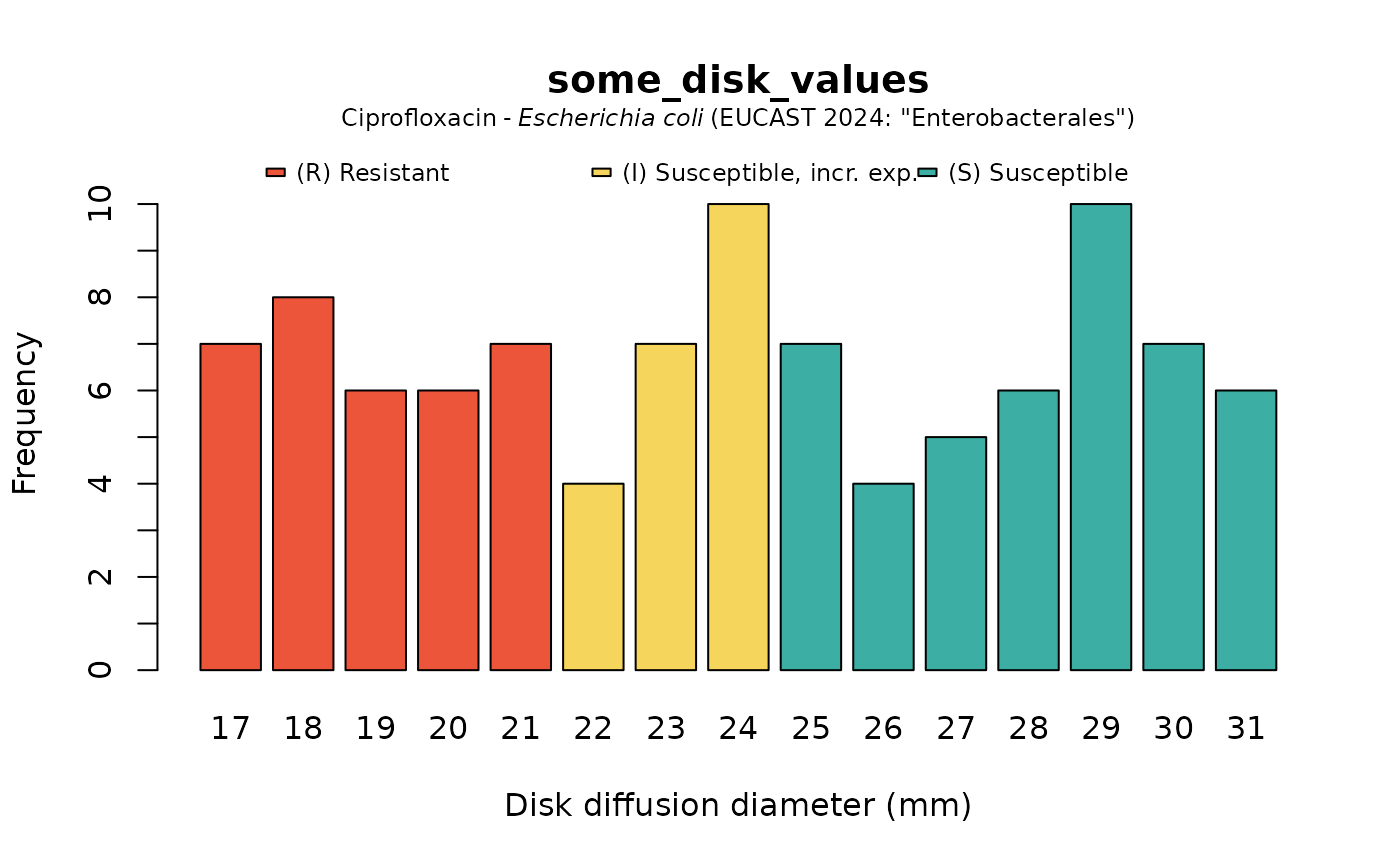
(image error) Size: 39 KiB After 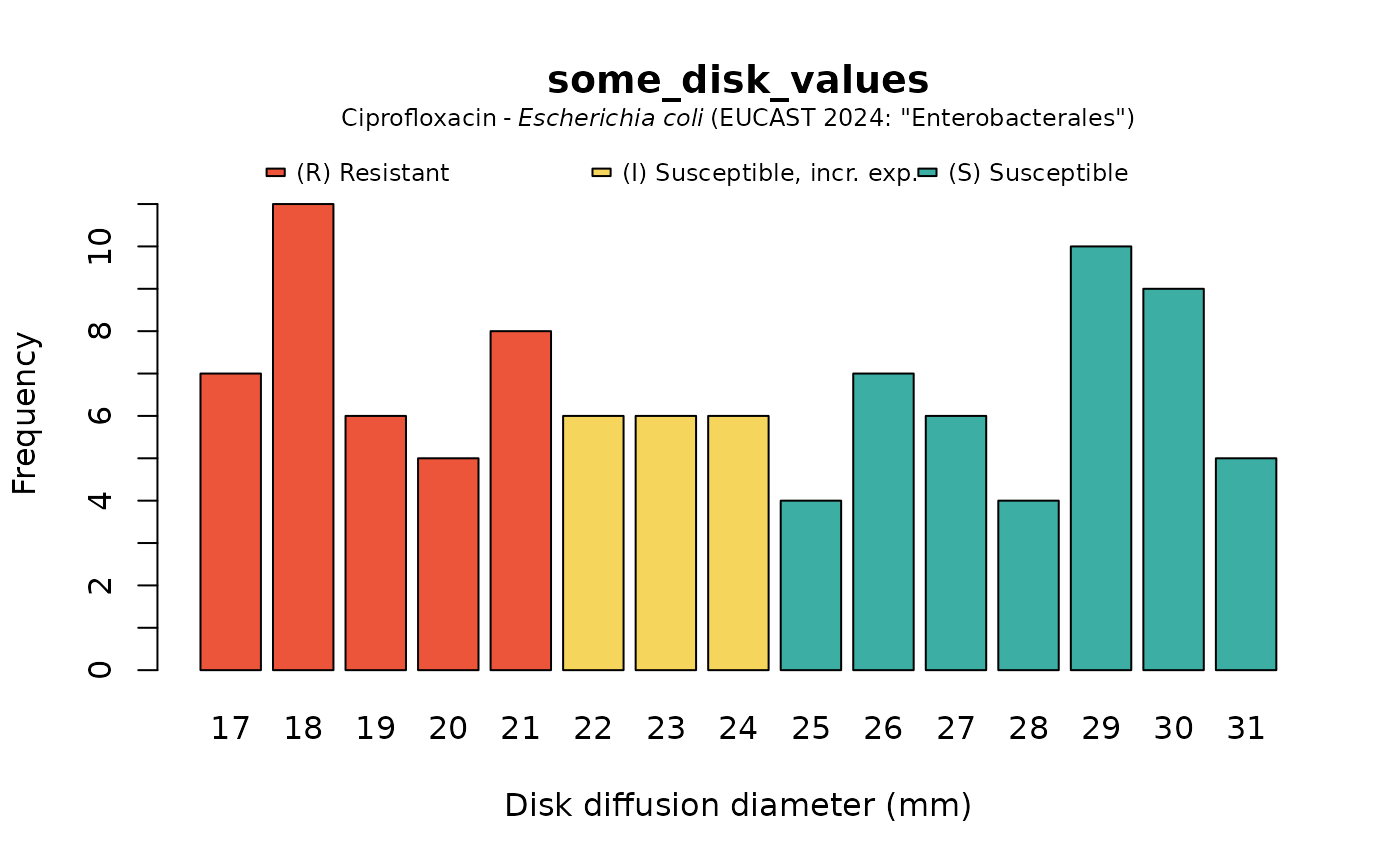
(image error) Size: 39 KiB 

|
|
Before 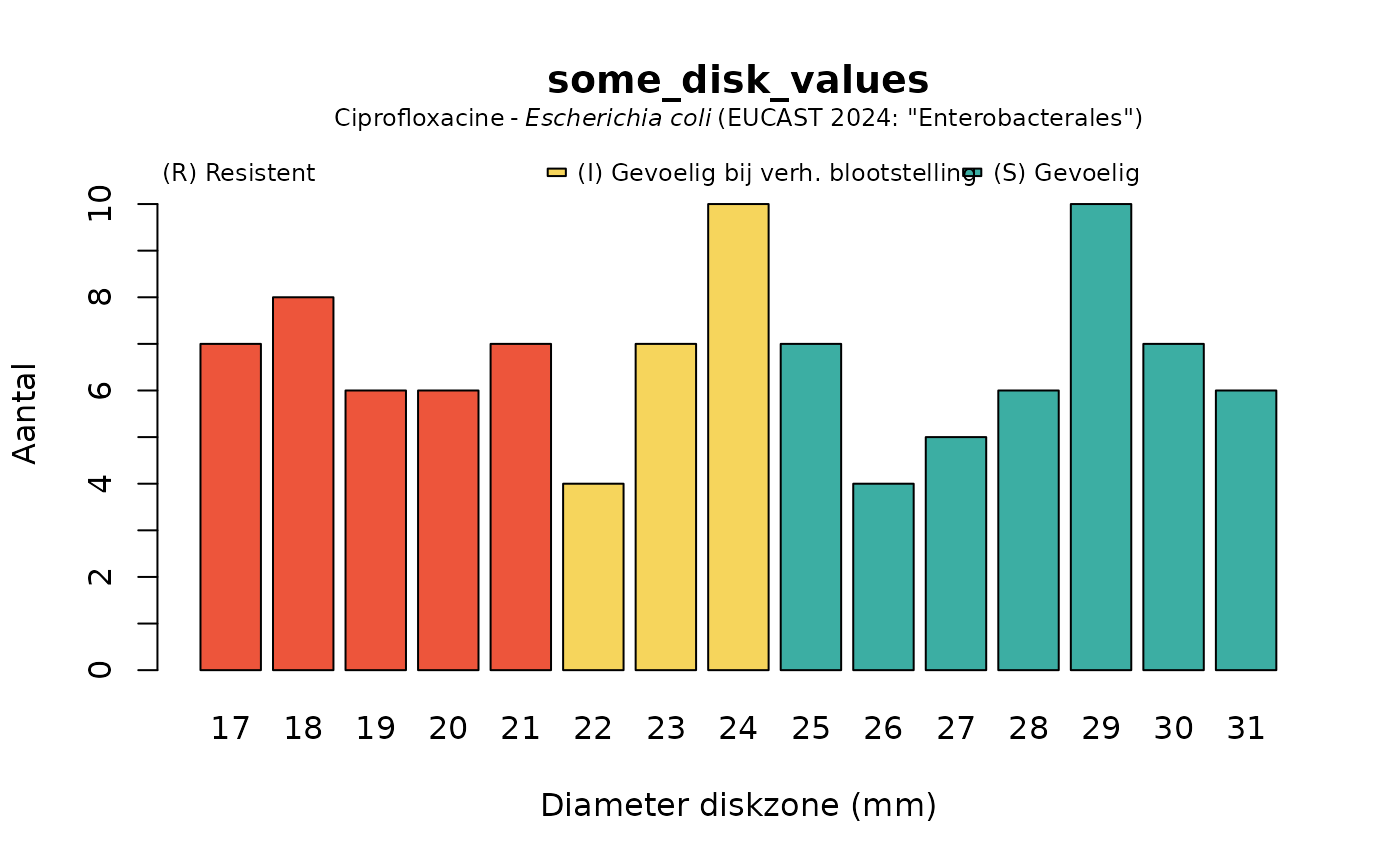
(image error) Size: 38 KiB After 
(image error) Size: 39 KiB 

|
@ -9,7 +9,7 @@ Especially the scale_*_mic() functions are relevant wrappers to plot MIC values
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -57,137 +57,92 @@ Especially the scale_*_mic() functions are relevant wrappers to plot MIC values
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">scale_x_mic</span><span class="op">(</span>keep_operators <span class="op">=</span> <span class="st">"edges"</span>, mic_range <span class="op">=</span> <span class="cn">NULL</span>, drop <span class="op">=</span> <span class="cn">FALSE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">scale_x_mic</span><span class="op">(</span>keep_operators <span class="op">=</span> <span class="st">"edges"</span>, mic_range <span class="op">=</span> <span class="cn">NULL</span>, drop <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">scale_y_mic</span><span class="op">(</span>keep_operators <span class="op">=</span> <span class="st">"edges"</span>, mic_range <span class="op">=</span> <span class="cn">NULL</span>, drop <span class="op">=</span> <span class="cn">FALSE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span><span class="fu">scale_y_mic</span><span class="op">(</span>keep_operators <span class="op">=</span> <span class="st">"edges"</span>, mic_range <span class="op">=</span> <span class="cn">NULL</span>, drop <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">scale_colour_mic</span><span class="op">(</span>keep_operators <span class="op">=</span> <span class="st">"edges"</span>, mic_range <span class="op">=</span> <span class="cn">NULL</span>, drop <span class="op">=</span> <span class="cn">FALSE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span><span class="fu">scale_colour_mic</span><span class="op">(</span>keep_operators <span class="op">=</span> <span class="st">"edges"</span>, mic_range <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> drop <span class="op">=</span> <span class="cn">FALSE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">scale_fill_mic</span><span class="op">(</span>keep_operators <span class="op">=</span> <span class="st">"edges"</span>, mic_range <span class="op">=</span> <span class="cn">NULL</span>, drop <span class="op">=</span> <span class="cn">FALSE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span><span class="fu">scale_fill_mic</span><span class="op">(</span>keep_operators <span class="op">=</span> <span class="st">"edges"</span>, mic_range <span class="op">=</span> <span class="cn">NULL</span>, drop <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="co"># S3 method for class 'mic'</span></span>
|
||||
<span><span class="fu">plot</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> mo <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> ab <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> guideline <span class="op">=</span> <span class="st">"EUCAST"</span>,</span>
|
||||
<span> main <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/deparse.html" class="external-link">deparse</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/substitute.html" class="external-link">substitute</a></span><span class="op">(</span><span class="va">x</span><span class="op">)</span><span class="op">)</span>,</span>
|
||||
<span> ylab <span class="op">=</span> <span class="fu"><a href="translate.html">translate_AMR</a></span><span class="op">(</span><span class="st">"Frequency"</span>, language <span class="op">=</span> <span class="va">language</span><span class="op">)</span>,</span>
|
||||
<span> xlab <span class="op">=</span> <span class="fu"><a href="translate.html">translate_AMR</a></span><span class="op">(</span><span class="st">"Minimum Inhibitory Concentration (mg/L)"</span>, language <span class="op">=</span> <span class="va">language</span><span class="op">)</span>,</span>
|
||||
<span> colours_SIR <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"#3CAEA3"</span>, <span class="st">"#F6D55C"</span>, <span class="st">"#ED553B"</span><span class="op">)</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> expand <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span><span class="fu">plot</span><span class="op">(</span><span class="va">x</span>, mo <span class="op">=</span> <span class="cn">NULL</span>, ab <span class="op">=</span> <span class="cn">NULL</span>, guideline <span class="op">=</span> <span class="st">"EUCAST"</span>,</span>
|
||||
<span> main <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/deparse.html" class="external-link">deparse</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/substitute.html" class="external-link">substitute</a></span><span class="op">(</span><span class="va">x</span><span class="op">)</span><span class="op">)</span>, ylab <span class="op">=</span> <span class="fu"><a href="translate.html">translate_AMR</a></span><span class="op">(</span><span class="st">"Frequency"</span>, language</span>
|
||||
<span> <span class="op">=</span> <span class="va">language</span><span class="op">)</span>,</span>
|
||||
<span> xlab <span class="op">=</span> <span class="fu"><a href="translate.html">translate_AMR</a></span><span class="op">(</span><span class="st">"Minimum Inhibitory Concentration (mg/L)"</span>, language <span class="op">=</span></span>
|
||||
<span> <span class="va">language</span><span class="op">)</span>, colours_SIR <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"#3CAEA3"</span>, <span class="st">"#F6D55C"</span>, <span class="st">"#ED553B"</span><span class="op">)</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>, expand <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> include_PKPD <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_include_PKPD"</span>, <span class="cn">TRUE</span><span class="op">)</span>,</span>
|
||||
<span> breakpoint_type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_breakpoint_type"</span>, <span class="st">"human"</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span> breakpoint_type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_breakpoint_type"</span>, <span class="st">"human"</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="co"># S3 method for class 'mic'</span></span>
|
||||
<span><span class="fu"><a href="https://ggplot2.tidyverse.org/reference/autoplot.html" class="external-link">autoplot</a></span><span class="op">(</span></span>
|
||||
<span> <span class="va">object</span>,</span>
|
||||
<span> mo <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> ab <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> guideline <span class="op">=</span> <span class="st">"EUCAST"</span>,</span>
|
||||
<span> title <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/deparse.html" class="external-link">deparse</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/substitute.html" class="external-link">substitute</a></span><span class="op">(</span><span class="va">object</span><span class="op">)</span><span class="op">)</span>,</span>
|
||||
<span><span class="fu"><a href="https://ggplot2.tidyverse.org/reference/autoplot.html" class="external-link">autoplot</a></span><span class="op">(</span><span class="va">object</span>, mo <span class="op">=</span> <span class="cn">NULL</span>, ab <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> guideline <span class="op">=</span> <span class="st">"EUCAST"</span>, title <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/deparse.html" class="external-link">deparse</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/substitute.html" class="external-link">substitute</a></span><span class="op">(</span><span class="va">object</span><span class="op">)</span><span class="op">)</span>,</span>
|
||||
<span> ylab <span class="op">=</span> <span class="fu"><a href="translate.html">translate_AMR</a></span><span class="op">(</span><span class="st">"Frequency"</span>, language <span class="op">=</span> <span class="va">language</span><span class="op">)</span>,</span>
|
||||
<span> xlab <span class="op">=</span> <span class="fu"><a href="translate.html">translate_AMR</a></span><span class="op">(</span><span class="st">"Minimum Inhibitory Concentration (mg/L)"</span>, language <span class="op">=</span> <span class="va">language</span><span class="op">)</span>,</span>
|
||||
<span> colours_SIR <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"#3CAEA3"</span>, <span class="st">"#F6D55C"</span>, <span class="st">"#ED553B"</span><span class="op">)</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> expand <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> xlab <span class="op">=</span> <span class="fu"><a href="translate.html">translate_AMR</a></span><span class="op">(</span><span class="st">"Minimum Inhibitory Concentration (mg/L)"</span>, language <span class="op">=</span></span>
|
||||
<span> <span class="va">language</span><span class="op">)</span>, colours_SIR <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"#3CAEA3"</span>, <span class="st">"#F6D55C"</span>, <span class="st">"#ED553B"</span><span class="op">)</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>, expand <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> include_PKPD <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_include_PKPD"</span>, <span class="cn">TRUE</span><span class="op">)</span>,</span>
|
||||
<span> breakpoint_type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_breakpoint_type"</span>, <span class="st">"human"</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span> breakpoint_type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_breakpoint_type"</span>, <span class="st">"human"</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="co"># S3 method for class 'mic'</span></span>
|
||||
<span><span class="fu"><a href="https://ggplot2.tidyverse.org/reference/fortify.html" class="external-link">fortify</a></span><span class="op">(</span><span class="va">object</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="co"># S3 method for class 'disk'</span></span>
|
||||
<span><span class="fu">plot</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> main <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/deparse.html" class="external-link">deparse</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/substitute.html" class="external-link">substitute</a></span><span class="op">(</span><span class="va">x</span><span class="op">)</span><span class="op">)</span>,</span>
|
||||
<span><span class="fu">plot</span><span class="op">(</span><span class="va">x</span>, main <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/deparse.html" class="external-link">deparse</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/substitute.html" class="external-link">substitute</a></span><span class="op">(</span><span class="va">x</span><span class="op">)</span><span class="op">)</span>,</span>
|
||||
<span> ylab <span class="op">=</span> <span class="fu"><a href="translate.html">translate_AMR</a></span><span class="op">(</span><span class="st">"Frequency"</span>, language <span class="op">=</span> <span class="va">language</span><span class="op">)</span>,</span>
|
||||
<span> xlab <span class="op">=</span> <span class="fu"><a href="translate.html">translate_AMR</a></span><span class="op">(</span><span class="st">"Disk diffusion diameter (mm)"</span>, language <span class="op">=</span> <span class="va">language</span><span class="op">)</span>,</span>
|
||||
<span> mo <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> ab <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> guideline <span class="op">=</span> <span class="st">"EUCAST"</span>,</span>
|
||||
<span> mo <span class="op">=</span> <span class="cn">NULL</span>, ab <span class="op">=</span> <span class="cn">NULL</span>, guideline <span class="op">=</span> <span class="st">"EUCAST"</span>,</span>
|
||||
<span> colours_SIR <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"#3CAEA3"</span>, <span class="st">"#F6D55C"</span>, <span class="st">"#ED553B"</span><span class="op">)</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> expand <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>, expand <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> include_PKPD <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_include_PKPD"</span>, <span class="cn">TRUE</span><span class="op">)</span>,</span>
|
||||
<span> breakpoint_type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_breakpoint_type"</span>, <span class="st">"human"</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span> breakpoint_type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_breakpoint_type"</span>, <span class="st">"human"</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="co"># S3 method for class 'disk'</span></span>
|
||||
<span><span class="fu"><a href="https://ggplot2.tidyverse.org/reference/autoplot.html" class="external-link">autoplot</a></span><span class="op">(</span></span>
|
||||
<span> <span class="va">object</span>,</span>
|
||||
<span> mo <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> ab <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> title <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/deparse.html" class="external-link">deparse</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/substitute.html" class="external-link">substitute</a></span><span class="op">(</span><span class="va">object</span><span class="op">)</span><span class="op">)</span>,</span>
|
||||
<span> ylab <span class="op">=</span> <span class="fu"><a href="translate.html">translate_AMR</a></span><span class="op">(</span><span class="st">"Frequency"</span>, language <span class="op">=</span> <span class="va">language</span><span class="op">)</span>,</span>
|
||||
<span> xlab <span class="op">=</span> <span class="fu"><a href="translate.html">translate_AMR</a></span><span class="op">(</span><span class="st">"Disk diffusion diameter (mm)"</span>, language <span class="op">=</span> <span class="va">language</span><span class="op">)</span>,</span>
|
||||
<span> guideline <span class="op">=</span> <span class="st">"EUCAST"</span>,</span>
|
||||
<span> colours_SIR <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"#3CAEA3"</span>, <span class="st">"#F6D55C"</span>, <span class="st">"#ED553B"</span><span class="op">)</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> expand <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span><span class="fu"><a href="https://ggplot2.tidyverse.org/reference/autoplot.html" class="external-link">autoplot</a></span><span class="op">(</span><span class="va">object</span>, mo <span class="op">=</span> <span class="cn">NULL</span>, ab <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> title <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/deparse.html" class="external-link">deparse</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/substitute.html" class="external-link">substitute</a></span><span class="op">(</span><span class="va">object</span><span class="op">)</span><span class="op">)</span>, ylab <span class="op">=</span> <span class="fu"><a href="translate.html">translate_AMR</a></span><span class="op">(</span><span class="st">"Frequency"</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="va">language</span><span class="op">)</span>, xlab <span class="op">=</span> <span class="fu"><a href="translate.html">translate_AMR</a></span><span class="op">(</span><span class="st">"Disk diffusion diameter (mm)"</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="va">language</span><span class="op">)</span>, guideline <span class="op">=</span> <span class="st">"EUCAST"</span>, colours_SIR <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"#3CAEA3"</span>,</span>
|
||||
<span> <span class="st">"#F6D55C"</span>, <span class="st">"#ED553B"</span><span class="op">)</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>, expand <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> include_PKPD <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_include_PKPD"</span>, <span class="cn">TRUE</span><span class="op">)</span>,</span>
|
||||
<span> breakpoint_type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_breakpoint_type"</span>, <span class="st">"human"</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span> breakpoint_type <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/options.html" class="external-link">getOption</a></span><span class="op">(</span><span class="st">"AMR_breakpoint_type"</span>, <span class="st">"human"</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="co"># S3 method for class 'disk'</span></span>
|
||||
<span><span class="fu"><a href="https://ggplot2.tidyverse.org/reference/fortify.html" class="external-link">fortify</a></span><span class="op">(</span><span class="va">object</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="co"># S3 method for class 'sir'</span></span>
|
||||
<span><span class="fu">plot</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> ylab <span class="op">=</span> <span class="fu"><a href="translate.html">translate_AMR</a></span><span class="op">(</span><span class="st">"Percentage"</span>, language <span class="op">=</span> <span class="va">language</span><span class="op">)</span>,</span>
|
||||
<span> xlab <span class="op">=</span> <span class="fu"><a href="translate.html">translate_AMR</a></span><span class="op">(</span><span class="st">"Antimicrobial Interpretation"</span>, language <span class="op">=</span> <span class="va">language</span><span class="op">)</span>,</span>
|
||||
<span> main <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/deparse.html" class="external-link">deparse</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/substitute.html" class="external-link">substitute</a></span><span class="op">(</span><span class="va">x</span><span class="op">)</span><span class="op">)</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">plot</span><span class="op">(</span><span class="va">x</span>, ylab <span class="op">=</span> <span class="fu"><a href="translate.html">translate_AMR</a></span><span class="op">(</span><span class="st">"Percentage"</span>, language <span class="op">=</span></span>
|
||||
<span> <span class="va">language</span><span class="op">)</span>, xlab <span class="op">=</span> <span class="fu"><a href="translate.html">translate_AMR</a></span><span class="op">(</span><span class="st">"Antimicrobial Interpretation"</span>, language <span class="op">=</span></span>
|
||||
<span> <span class="va">language</span><span class="op">)</span>, main <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/deparse.html" class="external-link">deparse</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/substitute.html" class="external-link">substitute</a></span><span class="op">(</span><span class="va">x</span><span class="op">)</span><span class="op">)</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="co"># S3 method for class 'sir'</span></span>
|
||||
<span><span class="fu"><a href="https://ggplot2.tidyverse.org/reference/autoplot.html" class="external-link">autoplot</a></span><span class="op">(</span></span>
|
||||
<span> <span class="va">object</span>,</span>
|
||||
<span> title <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/deparse.html" class="external-link">deparse</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/substitute.html" class="external-link">substitute</a></span><span class="op">(</span><span class="va">object</span><span class="op">)</span><span class="op">)</span>,</span>
|
||||
<span><span class="fu"><a href="https://ggplot2.tidyverse.org/reference/autoplot.html" class="external-link">autoplot</a></span><span class="op">(</span><span class="va">object</span>, title <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/deparse.html" class="external-link">deparse</a></span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/base/substitute.html" class="external-link">substitute</a></span><span class="op">(</span><span class="va">object</span><span class="op">)</span><span class="op">)</span>,</span>
|
||||
<span> xlab <span class="op">=</span> <span class="fu"><a href="translate.html">translate_AMR</a></span><span class="op">(</span><span class="st">"Antimicrobial Interpretation"</span>, language <span class="op">=</span> <span class="va">language</span><span class="op">)</span>,</span>
|
||||
<span> ylab <span class="op">=</span> <span class="fu"><a href="translate.html">translate_AMR</a></span><span class="op">(</span><span class="st">"Frequency"</span>, language <span class="op">=</span> <span class="va">language</span><span class="op">)</span>,</span>
|
||||
<span> colours_SIR <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"#3CAEA3"</span>, <span class="st">"#F6D55C"</span>, <span class="st">"#ED553B"</span><span class="op">)</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="co"># S3 method for class 'sir'</span></span>
|
||||
<span><span class="fu"><a href="https://ggplot2.tidyverse.org/reference/fortify.html" class="external-link">fortify</a></span><span class="op">(</span><span class="va">object</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">facet_sir</span><span class="op">(</span>facet <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"interpretation"</span>, <span class="st">"antibiotic"</span><span class="op">)</span>, nrow <span class="op">=</span> <span class="cn">NULL</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">scale_y_percent</span><span class="op">(</span></span>
|
||||
<span> breaks <span class="op">=</span> <span class="kw">function</span><span class="op">(</span><span class="va">x</span><span class="op">)</span> <span class="fu"><a href="https://rdrr.io/r/base/seq.html" class="external-link">seq</a></span><span class="op">(</span><span class="fl">0</span>, <span class="fu"><a href="https://rdrr.io/r/base/Extremes.html" class="external-link">max</a></span><span class="op">(</span><span class="va">x</span>, na.rm <span class="op">=</span> <span class="cn">TRUE</span><span class="op">)</span>, <span class="fl">0.1</span><span class="op">)</span>,</span>
|
||||
<span> limits <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="fl">0</span>, <span class="cn">NA</span><span class="op">)</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">scale_y_percent</span><span class="op">(</span>breaks <span class="op">=</span> <span class="kw">function</span><span class="op">(</span><span class="va">x</span><span class="op">)</span> <span class="fu"><a href="https://rdrr.io/r/base/seq.html" class="external-link">seq</a></span><span class="op">(</span><span class="fl">0</span>, <span class="fu"><a href="https://rdrr.io/r/base/Extremes.html" class="external-link">max</a></span><span class="op">(</span><span class="va">x</span>, na.rm <span class="op">=</span> <span class="cn">TRUE</span><span class="op">)</span>, <span class="fl">0.1</span><span class="op">)</span>,</span>
|
||||
<span> limits <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="fl">0</span>, <span class="cn">NA</span><span class="op">)</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">scale_sir_colours</span><span class="op">(</span></span>
|
||||
<span> <span class="va">...</span>,</span>
|
||||
<span> aesthetics <span class="op">=</span> <span class="st">"fill"</span>,</span>
|
||||
<span> colours_SIR <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"#3CAEA3"</span>, <span class="st">"#F6D55C"</span>, <span class="st">"#ED553B"</span><span class="op">)</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">scale_sir_colours</span><span class="op">(</span><span class="va">...</span>, aesthetics <span class="op">=</span> <span class="st">"fill"</span>, colours_SIR <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/c.html" class="external-link">c</a></span><span class="op">(</span><span class="st">"#3CAEA3"</span>,</span>
|
||||
<span> <span class="st">"#F6D55C"</span>, <span class="st">"#ED553B"</span><span class="op">)</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">theme_sir</span><span class="op">(</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">labels_sir_count</span><span class="op">(</span></span>
|
||||
<span> position <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> x <span class="op">=</span> <span class="st">"antibiotic"</span>,</span>
|
||||
<span> translate_ab <span class="op">=</span> <span class="st">"name"</span>,</span>
|
||||
<span> minimum <span class="op">=</span> <span class="fl">30</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> combine_SI <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> datalabels.size <span class="op">=</span> <span class="fl">3</span>,</span>
|
||||
<span> datalabels.colour <span class="op">=</span> <span class="st">"grey15"</span></span>
|
||||
<span><span class="op">)</span></span></code></pre></div>
|
||||
<span><span class="fu">labels_sir_count</span><span class="op">(</span>position <span class="op">=</span> <span class="cn">NULL</span>, x <span class="op">=</span> <span class="st">"antibiotic"</span>,</span>
|
||||
<span> translate_ab <span class="op">=</span> <span class="st">"name"</span>, minimum <span class="op">=</span> <span class="fl">30</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> combine_SI <span class="op">=</span> <span class="cn">TRUE</span>, datalabels.size <span class="op">=</span> <span class="fl">3</span>, datalabels.colour <span class="op">=</span> <span class="st">"grey15"</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
|
||||
@ -9,7 +9,7 @@ resistance() should be used to calculate resistance, susceptibility() should be
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -57,50 +57,38 @@ resistance() should be used to calculate resistance, susceptibility() should be
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">resistance</span><span class="op">(</span><span class="va">...</span>, minimum <span class="op">=</span> <span class="fl">30</span>, as_percent <span class="op">=</span> <span class="cn">FALSE</span>, only_all_tested <span class="op">=</span> <span class="cn">FALSE</span><span class="op">)</span></span>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">resistance</span><span class="op">(</span><span class="va">...</span>, minimum <span class="op">=</span> <span class="fl">30</span>, as_percent <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_all_tested <span class="op">=</span> <span class="cn">FALSE</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">susceptibility</span><span class="op">(</span><span class="va">...</span>, minimum <span class="op">=</span> <span class="fl">30</span>, as_percent <span class="op">=</span> <span class="cn">FALSE</span>, only_all_tested <span class="op">=</span> <span class="cn">FALSE</span><span class="op">)</span></span>
|
||||
<span><span class="fu">susceptibility</span><span class="op">(</span><span class="va">...</span>, minimum <span class="op">=</span> <span class="fl">30</span>, as_percent <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_all_tested <span class="op">=</span> <span class="cn">FALSE</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">sir_confidence_interval</span><span class="op">(</span></span>
|
||||
<span> <span class="va">...</span>,</span>
|
||||
<span> ab_result <span class="op">=</span> <span class="st">"R"</span>,</span>
|
||||
<span> minimum <span class="op">=</span> <span class="fl">30</span>,</span>
|
||||
<span> as_percent <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_all_tested <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> confidence_level <span class="op">=</span> <span class="fl">0.95</span>,</span>
|
||||
<span> side <span class="op">=</span> <span class="st">"both"</span>,</span>
|
||||
<span> collapse <span class="op">=</span> <span class="cn">FALSE</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">sir_confidence_interval</span><span class="op">(</span><span class="va">...</span>, ab_result <span class="op">=</span> <span class="st">"R"</span>, minimum <span class="op">=</span> <span class="fl">30</span>,</span>
|
||||
<span> as_percent <span class="op">=</span> <span class="cn">FALSE</span>, only_all_tested <span class="op">=</span> <span class="cn">FALSE</span>, confidence_level <span class="op">=</span> <span class="fl">0.95</span>,</span>
|
||||
<span> side <span class="op">=</span> <span class="st">"both"</span>, collapse <span class="op">=</span> <span class="cn">FALSE</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">proportion_R</span><span class="op">(</span><span class="va">...</span>, minimum <span class="op">=</span> <span class="fl">30</span>, as_percent <span class="op">=</span> <span class="cn">FALSE</span>, only_all_tested <span class="op">=</span> <span class="cn">FALSE</span><span class="op">)</span></span>
|
||||
<span><span class="fu">proportion_R</span><span class="op">(</span><span class="va">...</span>, minimum <span class="op">=</span> <span class="fl">30</span>, as_percent <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_all_tested <span class="op">=</span> <span class="cn">FALSE</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">proportion_IR</span><span class="op">(</span><span class="va">...</span>, minimum <span class="op">=</span> <span class="fl">30</span>, as_percent <span class="op">=</span> <span class="cn">FALSE</span>, only_all_tested <span class="op">=</span> <span class="cn">FALSE</span><span class="op">)</span></span>
|
||||
<span><span class="fu">proportion_IR</span><span class="op">(</span><span class="va">...</span>, minimum <span class="op">=</span> <span class="fl">30</span>, as_percent <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_all_tested <span class="op">=</span> <span class="cn">FALSE</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">proportion_I</span><span class="op">(</span><span class="va">...</span>, minimum <span class="op">=</span> <span class="fl">30</span>, as_percent <span class="op">=</span> <span class="cn">FALSE</span>, only_all_tested <span class="op">=</span> <span class="cn">FALSE</span><span class="op">)</span></span>
|
||||
<span><span class="fu">proportion_I</span><span class="op">(</span><span class="va">...</span>, minimum <span class="op">=</span> <span class="fl">30</span>, as_percent <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_all_tested <span class="op">=</span> <span class="cn">FALSE</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">proportion_SI</span><span class="op">(</span><span class="va">...</span>, minimum <span class="op">=</span> <span class="fl">30</span>, as_percent <span class="op">=</span> <span class="cn">FALSE</span>, only_all_tested <span class="op">=</span> <span class="cn">FALSE</span><span class="op">)</span></span>
|
||||
<span><span class="fu">proportion_SI</span><span class="op">(</span><span class="va">...</span>, minimum <span class="op">=</span> <span class="fl">30</span>, as_percent <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_all_tested <span class="op">=</span> <span class="cn">FALSE</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">proportion_S</span><span class="op">(</span><span class="va">...</span>, minimum <span class="op">=</span> <span class="fl">30</span>, as_percent <span class="op">=</span> <span class="cn">FALSE</span>, only_all_tested <span class="op">=</span> <span class="cn">FALSE</span><span class="op">)</span></span>
|
||||
<span><span class="fu">proportion_S</span><span class="op">(</span><span class="va">...</span>, minimum <span class="op">=</span> <span class="fl">30</span>, as_percent <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> only_all_tested <span class="op">=</span> <span class="cn">FALSE</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">proportion_df</span><span class="op">(</span></span>
|
||||
<span> <span class="va">data</span>,</span>
|
||||
<span> translate_ab <span class="op">=</span> <span class="st">"name"</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> minimum <span class="op">=</span> <span class="fl">30</span>,</span>
|
||||
<span> as_percent <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> combine_SI <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> confidence_level <span class="op">=</span> <span class="fl">0.95</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">proportion_df</span><span class="op">(</span><span class="va">data</span>, translate_ab <span class="op">=</span> <span class="st">"name"</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> minimum <span class="op">=</span> <span class="fl">30</span>, as_percent <span class="op">=</span> <span class="cn">FALSE</span>, combine_SI <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> confidence_level <span class="op">=</span> <span class="fl">0.95</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">sir_df</span><span class="op">(</span></span>
|
||||
<span> <span class="va">data</span>,</span>
|
||||
<span> translate_ab <span class="op">=</span> <span class="st">"name"</span>,</span>
|
||||
<span> language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> minimum <span class="op">=</span> <span class="fl">30</span>,</span>
|
||||
<span> as_percent <span class="op">=</span> <span class="cn">FALSE</span>,</span>
|
||||
<span> combine_SI <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> confidence_level <span class="op">=</span> <span class="fl">0.95</span></span>
|
||||
<span><span class="op">)</span></span></code></pre></div>
|
||||
<span><span class="fu">sir_df</span><span class="op">(</span><span class="va">data</span>, translate_ab <span class="op">=</span> <span class="st">"name"</span>, language <span class="op">=</span> <span class="fu"><a href="translate.html">get_AMR_locale</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> minimum <span class="op">=</span> <span class="fl">30</span>, as_percent <span class="op">=</span> <span class="cn">FALSE</span>, combine_SI <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> confidence_level <span class="op">=</span> <span class="fl">0.95</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -99,43 +99,43 @@
|
||||
<h2 id="ref-examples">Examples<a class="anchor" aria-label="anchor" href="#ref-examples"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span class="r-in"><span><span class="fu">random_mic</span><span class="op">(</span><span class="fl">25</span><span class="op">)</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> Class 'mic'</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 0.025 8 0.002 0.005 0.25 >=256 0.01 0.002 64 </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [10] 0.5 4 32 0.002 16 >=256 0.01 0.002 0.0625 </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [19] 0.002 64 8 8 2 <=0.001 0.125 </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 1 16 4 0.125 4 0.005 0.001 0.01 >=128 32 </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [11] 16 0.001 64 64 0.125 0.001 64 0.0625 0.002 0.005 </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [21] 64 2 0.25 0.01 1 </span>
|
||||
<span class="r-in"><span><span class="fu">random_disk</span><span class="op">(</span><span class="fl">25</span><span class="op">)</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> Class 'disk'</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 43 7 48 40 19 50 50 20 44 14 16 33 31 17 30 6 38 7 6 12 16 11 43 43 45</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 28 21 50 8 29 44 46 42 24 6 41 19 39 48 19 18 49 7 15 36 50 50 48 8 35</span>
|
||||
<span class="r-in"><span><span class="fu">random_sir</span><span class="op">(</span><span class="fl">25</span><span class="op">)</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> Class 'sir'</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] R S R R S S R I S R S S R I I R S I R R R R I R S</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] I I I I R S I I R I R S I R I I R R I S S I I S R</span>
|
||||
<span class="r-in"><span></span></span>
|
||||
<span class="r-in"><span><span class="co"># \donttest{</span></span></span>
|
||||
<span class="r-in"><span><span class="co"># make the random generation more realistic by setting a bug and/or drug:</span></span></span>
|
||||
<span class="r-in"><span><span class="fu">random_mic</span><span class="op">(</span><span class="fl">25</span>, <span class="st">"Klebsiella pneumoniae"</span><span class="op">)</span> <span class="co"># range 0.0625-64</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> Class 'mic'</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 0.025 0.25 16 8 0.001 4 16 0.025 0.25 8 </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [11] 0.5 0.005 0.0625 2 0.125 0.002 1 16 16 0.005 </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [21] 0.001 >=32 0.002 0.0625 0.125 </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 0.0625 16 0.025 4 32 0.025 0.002 0.0625 2 0.25 </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [11] 0.0625 0.125 0.001 16 0.001 1 0.25 0.5 64 0.005 </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [21] 4 4 0.125 0.002 0.5 </span>
|
||||
<span class="r-in"><span><span class="fu">random_mic</span><span class="op">(</span><span class="fl">25</span>, <span class="st">"Klebsiella pneumoniae"</span>, <span class="st">"meropenem"</span><span class="op">)</span> <span class="co"># range 0.0625-16</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> Class 'mic'</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 4 8 2 8 8 16 8 8 1 0.5 1 >=32 2 4 1 </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [16] >=32 4 2 4 4 2 16 4 16 2 </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] >=16 0.5 8 8 4 >=16 1 1 4 1 8 4 >=16 4 0.5 </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [16] 1 >=16 4 2 2 4 2 8 0.5 1 </span>
|
||||
<span class="r-in"><span><span class="fu">random_mic</span><span class="op">(</span><span class="fl">25</span>, <span class="st">"Streptococcus pneumoniae"</span>, <span class="st">"meropenem"</span><span class="op">)</span> <span class="co"># range 0.0625-4</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> Class 'mic'</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] <=0.0625 <=0.0625 <=0.0625 0.5 0.125 0.25 0.5 2 </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [9] 0.125 0.125 0.5 4 <=0.0625 2 0.25 4 </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [17] 2 0.25 1 2 2 <=0.0625 2 <=0.0625</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [25] 0.5 </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] >=8 4 2 <=0.0625 0.25 1 1 0.25 </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [9] <=0.0625 0.25 0.5 1 >=8 1 2 <=0.0625</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [17] >=8 <=0.0625 1 1 1 4 0.125 0.125 </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [25] 2 </span>
|
||||
<span class="r-in"><span></span></span>
|
||||
<span class="r-in"><span><span class="fu">random_disk</span><span class="op">(</span><span class="fl">25</span>, <span class="st">"Klebsiella pneumoniae"</span><span class="op">)</span> <span class="co"># range 8-50</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> Class 'disk'</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 30 43 22 49 40 8 14 36 49 23 12 9 17 25 43 46 30 49 34 22 50 24 39 38 46</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 15 46 20 39 24 36 50 42 15 12 11 17 40 16 46 44 48 50 24 50 29 14 39 43 14</span>
|
||||
<span class="r-in"><span><span class="fu">random_disk</span><span class="op">(</span><span class="fl">25</span>, <span class="st">"Klebsiella pneumoniae"</span>, <span class="st">"ampicillin"</span><span class="op">)</span> <span class="co"># range 11-17</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> Class 'disk'</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 17 15 16 14 17 14 12 13 13 17 17 12 14 16 15 14 16 14 16 13 11 16 17 13 13</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 16 11 11 13 11 16 17 16 11 11 14 11 11 14 16 14 16 12 12 13 14 13 17 12 11</span>
|
||||
<span class="r-in"><span><span class="fu">random_disk</span><span class="op">(</span><span class="fl">25</span>, <span class="st">"Streptococcus pneumoniae"</span>, <span class="st">"ampicillin"</span><span class="op">)</span> <span class="co"># range 12-27</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> Class 'disk'</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 25 22 21 23 19 20 21 25 20 19 21 19 27 22 26 25 27 21 17 26 18 23 23 26 27</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] 25 16 23 19 25 20 24 17 27 16 18 17 18 20 19 24 20 23 23 20 19 25 23 17 16</span>
|
||||
<span class="r-in"><span><span class="co"># }</span></span></span>
|
||||
</code></pre></div>
|
||||
</div>
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -54,53 +54,24 @@
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">resistance_predict</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> <span class="va">col_ab</span>,</span>
|
||||
<span> col_date <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> year_min <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> year_max <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> year_every <span class="op">=</span> <span class="fl">1</span>,</span>
|
||||
<span> minimum <span class="op">=</span> <span class="fl">30</span>,</span>
|
||||
<span> model <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> I_as_S <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> preserve_measurements <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> info <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/interactive.html" class="external-link">interactive</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">resistance_predict</span><span class="op">(</span><span class="va">x</span>, <span class="va">col_ab</span>, col_date <span class="op">=</span> <span class="cn">NULL</span>, year_min <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> year_max <span class="op">=</span> <span class="cn">NULL</span>, year_every <span class="op">=</span> <span class="fl">1</span>, minimum <span class="op">=</span> <span class="fl">30</span>, model <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> I_as_S <span class="op">=</span> <span class="cn">TRUE</span>, preserve_measurements <span class="op">=</span> <span class="cn">TRUE</span>, info <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/interactive.html" class="external-link">interactive</a></span><span class="op">(</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">sir_predict</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> <span class="va">col_ab</span>,</span>
|
||||
<span> col_date <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> year_min <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> year_max <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> year_every <span class="op">=</span> <span class="fl">1</span>,</span>
|
||||
<span> minimum <span class="op">=</span> <span class="fl">30</span>,</span>
|
||||
<span> model <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> I_as_S <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> preserve_measurements <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> info <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/interactive.html" class="external-link">interactive</a></span><span class="op">(</span><span class="op">)</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">sir_predict</span><span class="op">(</span><span class="va">x</span>, <span class="va">col_ab</span>, col_date <span class="op">=</span> <span class="cn">NULL</span>, year_min <span class="op">=</span> <span class="cn">NULL</span>, year_max <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> year_every <span class="op">=</span> <span class="fl">1</span>, minimum <span class="op">=</span> <span class="fl">30</span>, model <span class="op">=</span> <span class="cn">NULL</span>, I_as_S <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> preserve_measurements <span class="op">=</span> <span class="cn">TRUE</span>, info <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/interactive.html" class="external-link">interactive</a></span><span class="op">(</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="co"># S3 method for class 'resistance_predict'</span></span>
|
||||
<span><span class="fu"><a href="plot.html">plot</a></span><span class="op">(</span><span class="va">x</span>, main <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/paste.html" class="external-link">paste</a></span><span class="op">(</span><span class="st">"Resistance Prediction of"</span>, <span class="va">x_name</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span><span class="fu"><a href="plot.html">plot</a></span><span class="op">(</span><span class="va">x</span>, main <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/paste.html" class="external-link">paste</a></span><span class="op">(</span><span class="st">"Resistance Prediction of"</span>,</span>
|
||||
<span> <span class="va">x_name</span><span class="op">)</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="fu">ggplot_sir_predict</span><span class="op">(</span></span>
|
||||
<span> <span class="va">x</span>,</span>
|
||||
<span> main <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/paste.html" class="external-link">paste</a></span><span class="op">(</span><span class="st">"Resistance Prediction of"</span>, <span class="va">x_name</span><span class="op">)</span>,</span>
|
||||
<span> ribbon <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span>
|
||||
<span><span class="fu">ggplot_sir_predict</span><span class="op">(</span><span class="va">x</span>, main <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/paste.html" class="external-link">paste</a></span><span class="op">(</span><span class="st">"Resistance Prediction of"</span>, <span class="va">x_name</span><span class="op">)</span>,</span>
|
||||
<span> ribbon <span class="op">=</span> <span class="cn">TRUE</span>, <span class="va">...</span><span class="op">)</span></span>
|
||||
<span></span>
|
||||
<span><span class="co"># S3 method for class 'resistance_predict'</span></span>
|
||||
<span><span class="fu"><a href="https://ggplot2.tidyverse.org/reference/autoplot.html" class="external-link">autoplot</a></span><span class="op">(</span></span>
|
||||
<span> <span class="va">object</span>,</span>
|
||||
<span> main <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/paste.html" class="external-link">paste</a></span><span class="op">(</span><span class="st">"Resistance Prediction of"</span>, <span class="va">x_name</span><span class="op">)</span>,</span>
|
||||
<span> ribbon <span class="op">=</span> <span class="cn">TRUE</span>,</span>
|
||||
<span> <span class="va">...</span></span>
|
||||
<span><span class="op">)</span></span></code></pre></div>
|
||||
<span><span class="fu"><a href="https://ggplot2.tidyverse.org/reference/autoplot.html" class="external-link">autoplot</a></span><span class="op">(</span><span class="va">object</span>,</span>
|
||||
<span> main <span class="op">=</span> <span class="fu"><a href="https://rdrr.io/r/base/paste.html" class="external-link">paste</a></span><span class="op">(</span><span class="st">"Resistance Prediction of"</span>, <span class="va">x_name</span><span class="op">)</span>, ribbon <span class="op">=</span> <span class="cn">TRUE</span>, <span class="va">...</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
|
||||
@ -9,7 +9,7 @@ When negative ('left-skewed'): the left tail is longer; the mass of the distribu
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
@ -89,7 +89,7 @@ When negative ('left-skewed'): the left tail is longer; the mass of the distribu
|
||||
<div class="section level2">
|
||||
<h2 id="ref-examples">Examples<a class="anchor" aria-label="anchor" href="#ref-examples"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span class="r-in"><span><span class="fu">skewness</span><span class="op">(</span><span class="fu"><a href="https://rdrr.io/r/stats/Uniform.html" class="external-link">runif</a></span><span class="op">(</span><span class="fl">1000</span><span class="op">)</span><span class="op">)</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] -0.07394966</span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> [1] -0.02093725</span>
|
||||
</code></pre></div>
|
||||
</div>
|
||||
</main><aside class="col-md-3"><nav id="toc" aria-label="Table of contents"><h2>On this page</h2>
|
||||
|
||||
192
reference/top_n_microorganisms.html
Normal file
@ -0,0 +1,192 @@
|
||||
<!DOCTYPE html>
|
||||
<!-- Generated by pkgdown: do not edit by hand --><html lang="en"><head><meta http-equiv="Content-Type" content="text/html; charset=UTF-8"><meta charset="utf-8"><meta http-equiv="X-UA-Compatible" content="IE=edge"><meta name="viewport" content="width=device-width, initial-scale=1, shrink-to-fit=no"><title>Filter Top n Microorganisms — top_n_microorganisms • AMR (for R)</title><!-- favicons --><link rel="icon" type="image/png" sizes="16x16" href="../favicon-16x16.png"><link rel="icon" type="image/png" sizes="32x32" href="../favicon-32x32.png"><link rel="apple-touch-icon" type="image/png" sizes="180x180" href="../apple-touch-icon.png"><link rel="apple-touch-icon" type="image/png" sizes="120x120" href="../apple-touch-icon-120x120.png"><link rel="apple-touch-icon" type="image/png" sizes="76x76" href="../apple-touch-icon-76x76.png"><link rel="apple-touch-icon" type="image/png" sizes="60x60" href="../apple-touch-icon-60x60.png"><script src="../deps/jquery-3.6.0/jquery-3.6.0.min.js"></script><meta name="viewport" content="width=device-width, initial-scale=1, shrink-to-fit=no"><link href="../deps/bootstrap-5.3.1/bootstrap.min.css" rel="stylesheet"><script src="../deps/bootstrap-5.3.1/bootstrap.bundle.min.js"></script><link href="../deps/Lato-0.4.9/font.css" rel="stylesheet"><link href="../deps/Fira_Code-0.4.9/font.css" rel="stylesheet"><link href="../deps/font-awesome-6.5.2/css/all.min.css" rel="stylesheet"><link href="../deps/font-awesome-6.5.2/css/v4-shims.min.css" rel="stylesheet"><script src="../deps/headroom-0.11.0/headroom.min.js"></script><script src="../deps/headroom-0.11.0/jQuery.headroom.min.js"></script><script src="../deps/bootstrap-toc-1.0.1/bootstrap-toc.min.js"></script><script src="../deps/clipboard.js-2.0.11/clipboard.min.js"></script><script src="../deps/search-1.0.0/autocomplete.jquery.min.js"></script><script src="../deps/search-1.0.0/fuse.min.js"></script><script src="../deps/search-1.0.0/mark.min.js"></script><!-- pkgdown --><script src="../pkgdown.js"></script><link href="../extra.css" rel="stylesheet"><script src="../extra.js"></script><meta property="og:title" content="Filter Top n Microorganisms — top_n_microorganisms"><meta name="description" content="This function filters a data set to include only the top n microorganisms based on a specified property, such as taxonomic family or genus. For example, it can filter a data set to the top 3 species, or to any species in the top 5 genera, or to the top 3 species in each of the top 5 genera."><meta property="og:description" content="This function filters a data set to include only the top n microorganisms based on a specified property, such as taxonomic family or genus. For example, it can filter a data set to the top 3 species, or to any species in the top 5 genera, or to the top 3 species in each of the top 5 genera."><meta property="og:image" content="https://msberends.github.io/AMR/logo.svg"></head><body>
|
||||
<a href="#main" class="visually-hidden-focusable">Skip to contents</a>
|
||||
|
||||
|
||||
<nav class="navbar navbar-expand-lg fixed-top bg-primary" data-bs-theme="dark" aria-label="Site navigation"><div class="container">
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
<span class="navbar-toggler-icon"></span>
|
||||
</button>
|
||||
|
||||
<div id="navbar" class="collapse navbar-collapse ms-3">
|
||||
<ul class="navbar-nav me-auto"><li class="nav-item dropdown">
|
||||
<button class="nav-link dropdown-toggle" type="button" id="dropdown-how-to" data-bs-toggle="dropdown" aria-expanded="false" aria-haspopup="true"><span class="fa fa-question-circle"></span> How to</button>
|
||||
<ul class="dropdown-menu" aria-labelledby="dropdown-how-to"><li><a class="dropdown-item" href="../articles/AMR.html"><span class="fa fa-directions"></span> Conduct AMR Analysis</a></li>
|
||||
<li><a class="dropdown-item" href="../reference/antibiogram.html"><span class="fa fa-file-prescription"></span> Generate Antibiogram (Trad./Syndromic/WISCA)</a></li>
|
||||
<li><a class="dropdown-item" href="../articles/resistance_predict.html"><span class="fa fa-dice"></span> Predict Antimicrobial Resistance</a></li>
|
||||
<li><a class="dropdown-item" href="../articles/datasets.html"><span class="fa fa-database"></span> Download Data Sets for Own Use</a></li>
|
||||
<li><a class="dropdown-item" href="../articles/AMR_with_tidymodels.html"><span class="fa fa-square-root-variable"></span> Use AMR for Predictive Modelling (tidymodels)</a></li>
|
||||
<li><a class="dropdown-item" href="../reference/AMR-options.html"><span class="fa fa-gear"></span> Set User- Or Team-specific Package Settings</a></li>
|
||||
<li><a class="dropdown-item" href="../articles/PCA.html"><span class="fa fa-compress"></span> Conduct Principal Component Analysis for AMR</a></li>
|
||||
<li><a class="dropdown-item" href="../articles/MDR.html"><span class="fa fa-skull-crossbones"></span> Determine Multi-Drug Resistance (MDR)</a></li>
|
||||
<li><a class="dropdown-item" href="../articles/WHONET.html"><span class="fa fa-globe-americas"></span> Work with WHONET Data</a></li>
|
||||
<li><a class="dropdown-item" href="../articles/EUCAST.html"><span class="fa fa-exchange-alt"></span> Apply Eucast Rules</a></li>
|
||||
<li><a class="dropdown-item" href="../reference/mo_property.html"><span class="fa fa-bug"></span> Get Taxonomy of a Microorganism</a></li>
|
||||
<li><a class="dropdown-item" href="../reference/ab_property.html"><span class="fa fa-capsules"></span> Get Properties of an Antibiotic Drug</a></li>
|
||||
<li><a class="dropdown-item" href="../reference/av_property.html"><span class="fa fa-capsules"></span> Get Properties of an Antiviral Drug</a></li>
|
||||
</ul></li>
|
||||
<li class="nav-item"><a class="nav-link" href="../articles/AMR_for_Python.html"><span class="fa fab fa-python"></span> AMR for Python</a></li>
|
||||
<li class="active nav-item"><a class="nav-link" href="../reference/index.html"><span class="fa fa-book-open"></span> Manual</a></li>
|
||||
<li class="nav-item"><a class="nav-link" href="../authors.html"><span class="fa fa-users"></span> Authors</a></li>
|
||||
</ul><ul class="navbar-nav"><li class="nav-item"><a class="nav-link" href="../news/index.html"><span class="fa far fa-newspaper"></span> Changelog</a></li>
|
||||
<li class="nav-item"><a class="external-link nav-link" href="https://github.com/msberends/AMR"><span class="fa fab fa-github"></span> Source Code</a></li>
|
||||
</ul></div>
|
||||
|
||||
|
||||
</div>
|
||||
</nav><div class="container template-reference-topic">
|
||||
<div class="row">
|
||||
<main id="main" class="col-md-9"><div class="page-header">
|
||||
<img src="../logo.svg" class="logo" alt=""><h1>Filter Top <em>n</em> Microorganisms</h1>
|
||||
<small class="dont-index">Source: <a href="https://github.com/msberends/AMR/blob/main/R/top_n_microorganisms.R" class="external-link"><code>R/top_n_microorganisms.R</code></a></small>
|
||||
<div class="d-none name"><code>top_n_microorganisms.Rd</code></div>
|
||||
</div>
|
||||
|
||||
<div class="ref-description section level2">
|
||||
<p>This function filters a data set to include only the top <em>n</em> microorganisms based on a specified property, such as taxonomic family or genus. For example, it can filter a data set to the top 3 species, or to any species in the top 5 genera, or to the top 3 species in each of the top 5 genera.</p>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-usage">Usage<a class="anchor" aria-label="anchor" href="#ref-usage"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span><span class="fu">top_n_microorganisms</span><span class="op">(</span><span class="va">x</span>, <span class="va">n</span>, property <span class="op">=</span> <span class="st">"fullname"</span>, n_for_each <span class="op">=</span> <span class="cn">NULL</span>,</span>
|
||||
<span> col_mo <span class="op">=</span> <span class="cn">NULL</span>, <span class="va">...</span><span class="op">)</span></span></code></pre></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="arguments">Arguments<a class="anchor" aria-label="anchor" href="#arguments"></a></h2>
|
||||
|
||||
|
||||
<dl><dt id="arg-x">x<a class="anchor" aria-label="anchor" href="#arg-x"></a></dt>
|
||||
<dd><p>a data frame containing microbial data</p></dd>
|
||||
|
||||
|
||||
<dt id="arg-n">n<a class="anchor" aria-label="anchor" href="#arg-n"></a></dt>
|
||||
<dd><p>an integer specifying the maximum number of unique values of the <code>property</code> to include in the output</p></dd>
|
||||
|
||||
|
||||
<dt id="arg-property">property<a class="anchor" aria-label="anchor" href="#arg-property"></a></dt>
|
||||
<dd><p>a character string indicating the microorganism property to use for filtering. Must be one of the column names of the <a href="microorganisms.html">microorganisms</a> data set: "mo", "fullname", "status", "kingdom", "phylum", "class", "order", "family", "genus", "species", "subspecies", "rank", "ref", "oxygen_tolerance", "source", "lpsn", "lpsn_parent", "lpsn_renamed_to", "mycobank", "mycobank_parent", "mycobank_renamed_to", "gbif", "gbif_parent", "gbif_renamed_to", "prevalence", or "snomed". If <code>NULL</code>, the raw values from <code>col_mo</code> will be used without transformation.</p></dd>
|
||||
|
||||
|
||||
<dt id="arg-n-for-each">n_for_each<a class="anchor" aria-label="anchor" href="#arg-n-for-each"></a></dt>
|
||||
<dd><p>an optional integer specifying the maximum number of rows to retain for each value of the selected property. If <code>NULL</code>, all rows within the top <em>n</em> groups will be included.</p></dd>
|
||||
|
||||
|
||||
<dt id="arg-col-mo">col_mo<a class="anchor" aria-label="anchor" href="#arg-col-mo"></a></dt>
|
||||
<dd><p>A character string indicating the column in <code>x</code> that contains microorganism names or codes. Defaults to the first column of class <code><a href="as.mo.html">mo</a></code>. Values will be coerced using <code><a href="as.mo.html">as.mo()</a></code>.</p></dd>
|
||||
|
||||
|
||||
<dt id="arg--">...<a class="anchor" aria-label="anchor" href="#arg--"></a></dt>
|
||||
<dd><p>Additional arguments passed on to <code><a href="mo_property.html">mo_property()</a></code> when <code>property</code> is not <code>NULL</code>.</p></dd>
|
||||
|
||||
</dl></div>
|
||||
<div class="section level2">
|
||||
<h2 id="details">Details<a class="anchor" aria-label="anchor" href="#details"></a></h2>
|
||||
<p>This function is useful for preprocessing data before creating <a href="antibiogram.html">antibiograms</a> or other analyses that require focused subsets of microbial data. For example, it can filter a data set to only include isolates from the top 10 species.</p>
|
||||
</div>
|
||||
<div class="section level2">
|
||||
<h2 id="see-also">See also<a class="anchor" aria-label="anchor" href="#see-also"></a></h2>
|
||||
<div class="dont-index"><p><code><a href="mo_property.html">mo_property()</a></code>, <code><a href="as.mo.html">as.mo()</a></code>, <code><a href="antibiogram.html">antibiogram()</a></code></p></div>
|
||||
</div>
|
||||
|
||||
<div class="section level2">
|
||||
<h2 id="ref-examples">Examples<a class="anchor" aria-label="anchor" href="#ref-examples"></a></h2>
|
||||
<div class="sourceCode"><pre class="sourceCode r"><code><span class="r-in"><span><span class="co"># filter to the top 3 species:</span></span></span>
|
||||
<span class="r-in"><span><span class="fu">top_n_microorganisms</span><span class="op">(</span><span class="va">example_isolates</span>,</span></span>
|
||||
<span class="r-in"><span> n <span class="op">=</span> <span class="fl">3</span><span class="op">)</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># A tibble: 1,015 × 46</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> date patient age gender ward mo PEN OXA FLC AMX </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494; font-style: italic;"><date></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><dbl></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><mo></span> <span style="color: #949494; font-style: italic;"><sir></span> <span style="color: #949494; font-style: italic;"><sir></span> <span style="color: #949494; font-style: italic;"><sir></span> <span style="color: #949494; font-style: italic;"><sir></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 1</span> 2002-01-02 A77334 65 F Clinical B_ESCHR_COLI R NA NA NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 2</span> 2002-01-03 A77334 65 F Clinical B_ESCHR_COLI R NA NA NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 3</span> 2002-01-14 462729 78 M Clinical B_STPHY_AURS R NA S R </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 4</span> 2002-01-14 462729 78 M Clinical B_STPHY_AURS R NA S R </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 5</span> 2002-01-19 738003 71 M Clinical B_ESCHR_COLI R NA NA NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 6</span> 2002-01-19 738003 71 M Clinical B_ESCHR_COLI R NA NA NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 7</span> 2002-02-03 481442 76 M ICU B_STPHY_CONS R NA S NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 8</span> 2002-02-14 067927 45 F ICU B_STPHY_CONS R NA R NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 9</span> 2002-02-14 067927 45 F ICU B_STPHY_CONS S NA S NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">10</span> 2002-02-21 A56499 64 M Clinical B_STPHY_CONS S NA S NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># ℹ 1,005 more rows</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># ℹ 36 more variables: AMC <sir>, AMP <sir>, TZP <sir>, CZO <sir>, FEP <sir>,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># CXM <sir>, FOX <sir>, CTX <sir>, CAZ <sir>, CRO <sir>, GEN <sir>,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># TOB <sir>, AMK <sir>, KAN <sir>, TMP <sir>, SXT <sir>, NIT <sir>,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># FOS <sir>, LNZ <sir>, CIP <sir>, MFX <sir>, VAN <sir>, TEC <sir>,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># TCY <sir>, TGC <sir>, DOX <sir>, ERY <sir>, CLI <sir>, AZM <sir>,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># IPM <sir>, MEM <sir>, MTR <sir>, CHL <sir>, COL <sir>, MUP <sir>, …</span></span>
|
||||
<span class="r-in"><span></span></span>
|
||||
<span class="r-in"><span><span class="co"># filter to any species in the top 5 genera:</span></span></span>
|
||||
<span class="r-in"><span><span class="fu">top_n_microorganisms</span><span class="op">(</span><span class="va">example_isolates</span>,</span></span>
|
||||
<span class="r-in"><span> n <span class="op">=</span> <span class="fl">5</span>, property <span class="op">=</span> <span class="st">"genus"</span><span class="op">)</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># A tibble: 1,742 × 46</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> date patient age gender ward mo PEN OXA FLC AMX </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494; font-style: italic;"><date></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><dbl></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><mo></span> <span style="color: #949494; font-style: italic;"><sir></span> <span style="color: #949494; font-style: italic;"><sir></span> <span style="color: #949494; font-style: italic;"><sir></span> <span style="color: #949494; font-style: italic;"><sir></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 1</span> 2002-01-02 A77334 65 F Clinical B_ESCHR_COLI R NA NA NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 2</span> 2002-01-03 A77334 65 F Clinical B_ESCHR_COLI R NA NA NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 3</span> 2002-01-07 067927 45 F ICU B_STPHY_EPDR R NA R NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 4</span> 2002-01-07 067927 45 F ICU B_STPHY_EPDR R NA R NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 5</span> 2002-01-13 067927 45 F ICU B_STPHY_EPDR R NA R NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 6</span> 2002-01-13 067927 45 F ICU B_STPHY_EPDR R NA R NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 7</span> 2002-01-14 462729 78 M Clinical B_STPHY_AURS R NA S R </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 8</span> 2002-01-14 462729 78 M Clinical B_STPHY_AURS R NA S R </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 9</span> 2002-01-16 067927 45 F ICU B_STPHY_EPDR R NA R NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">10</span> 2002-01-17 858515 79 F ICU B_STPHY_EPDR R NA S NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># ℹ 1,732 more rows</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># ℹ 36 more variables: AMC <sir>, AMP <sir>, TZP <sir>, CZO <sir>, FEP <sir>,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># CXM <sir>, FOX <sir>, CTX <sir>, CAZ <sir>, CRO <sir>, GEN <sir>,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># TOB <sir>, AMK <sir>, KAN <sir>, TMP <sir>, SXT <sir>, NIT <sir>,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># FOS <sir>, LNZ <sir>, CIP <sir>, MFX <sir>, VAN <sir>, TEC <sir>,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># TCY <sir>, TGC <sir>, DOX <sir>, ERY <sir>, CLI <sir>, AZM <sir>,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># IPM <sir>, MEM <sir>, MTR <sir>, CHL <sir>, COL <sir>, MUP <sir>, …</span></span>
|
||||
<span class="r-in"><span></span></span>
|
||||
<span class="r-in"><span><span class="co"># filter to the top 3 species in each of the top 5 genera:</span></span></span>
|
||||
<span class="r-in"><span><span class="fu">top_n_microorganisms</span><span class="op">(</span><span class="va">example_isolates</span>,</span></span>
|
||||
<span class="r-in"><span> n <span class="op">=</span> <span class="fl">5</span>, property <span class="op">=</span> <span class="st">"genus"</span>, n_for_each <span class="op">=</span> <span class="fl">3</span><span class="op">)</span></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># A tibble: 1,497 × 46</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> date patient age gender ward mo PEN OXA FLC AMX </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494; font-style: italic;"><date></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><dbl></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><chr></span> <span style="color: #949494; font-style: italic;"><mo></span> <span style="color: #949494; font-style: italic;"><sir></span> <span style="color: #949494; font-style: italic;"><sir></span> <span style="color: #949494; font-style: italic;"><sir></span> <span style="color: #949494; font-style: italic;"><sir></span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 1</span> 2002-02-21 4FC193 69 M Clinical B_ENTRC_FACM NA NA NA NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 2</span> 2002-04-08 130252 78 M ICU B_ENTRC_FCLS NA NA NA NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 3</span> 2002-06-23 798871 82 M Clinical B_ENTRC_FCLS NA NA NA NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 4</span> 2002-06-23 798871 82 M Clinical B_ENTRC_FCLS NA NA NA NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 5</span> 2003-04-20 6BC362 62 M ICU B_ENTRC NA NA NA NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 6</span> 2003-04-21 6BC362 62 M ICU B_ENTRC NA NA NA NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 7</span> 2003-08-13 F35553 52 M ICU B_ENTRC_FCLS NA NA NA NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 8</span> 2003-09-05 F35553 52 M ICU B_ENTRC NA NA NA NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;"> 9</span> 2003-09-05 F35553 52 M ICU B_ENTRC_FCLS NA NA NA NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #BCBCBC;">10</span> 2003-09-28 1B0933 80 M Clinical B_ENTRC NA NA NA NA </span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># ℹ 1,487 more rows</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># ℹ 36 more variables: AMC <sir>, AMP <sir>, TZP <sir>, CZO <sir>, FEP <sir>,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># CXM <sir>, FOX <sir>, CTX <sir>, CAZ <sir>, CRO <sir>, GEN <sir>,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># TOB <sir>, AMK <sir>, KAN <sir>, TMP <sir>, SXT <sir>, NIT <sir>,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># FOS <sir>, LNZ <sir>, CIP <sir>, MFX <sir>, VAN <sir>, TEC <sir>,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># TCY <sir>, TGC <sir>, DOX <sir>, ERY <sir>, CLI <sir>, AZM <sir>,</span></span>
|
||||
<span class="r-out co"><span class="r-pr">#></span> <span style="color: #949494;"># IPM <sir>, MEM <sir>, MTR <sir>, CHL <sir>, COL <sir>, MUP <sir>, …</span></span>
|
||||
</code></pre></div>
|
||||
</div>
|
||||
</main><aside class="col-md-3"><nav id="toc" aria-label="Table of contents"><h2>On this page</h2>
|
||||
</nav></aside></div>
|
||||
|
||||
|
||||
<footer><div class="pkgdown-footer-left">
|
||||
<p><code>AMR</code> (for R). Free and open-source, licenced under the <a target="_blank" href="https://github.com/msberends/AMR/blob/main/LICENSE" class="external-link">GNU General Public License version 2.0 (GPL-2)</a>.<br>Developed at the <a target="_blank" href="https://www.rug.nl" class="external-link">University of Groningen</a> and <a target="_blank" href="https://www.umcg.nl" class="external-link">University Medical Center Groningen</a> in The Netherlands.</p>
|
||||
</div>
|
||||
|
||||
<div class="pkgdown-footer-right">
|
||||
<p><a target="_blank" href="https://www.rug.nl" class="external-link"><img src="https://github.com/msberends/AMR/raw/main/pkgdown/assets/logo_rug.svg" style="max-width: 150px;"></a><a target="_blank" href="https://www.umcg.nl" class="external-link"><img src="https://github.com/msberends/AMR/raw/main/pkgdown/assets/logo_umcg.svg" style="max-width: 150px;"></a></p>
|
||||
</div>
|
||||
|
||||
</footer></div>
|
||||
|
||||
|
||||
|
||||
|
||||
|
||||
</body></html>
|
||||
|
||||
@ -7,7 +7,7 @@
|
||||
|
||||
<a class="navbar-brand me-2" href="../index.html">AMR (for R)</a>
|
||||
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9125</small>
|
||||
<small class="nav-text text-muted me-auto" data-bs-toggle="tooltip" data-bs-placement="bottom" title="">2.1.1.9133</small>
|
||||
|
||||
|
||||
<button class="navbar-toggler" type="button" data-bs-toggle="collapse" data-bs-target="#navbar" aria-controls="navbar" aria-expanded="false" aria-label="Toggle navigation">
|
||||
|
||||
8
reference/wisca.html
Normal file
@ -0,0 +1,8 @@
|
||||
<html>
|
||||
<head>
|
||||
<meta http-equiv="refresh" content="0;URL=https://msberends.github.io/AMR/reference/antibiogram.html" />
|
||||
<meta name="robots" content="noindex">
|
||||
<link rel="canonical" href="https://msberends.github.io/AMR/reference/antibiogram.html">
|
||||
</head>
|
||||
</html>
|
||||
|
||||
@ -75,6 +75,7 @@
|
||||
<url><loc>https://msberends.github.io/AMR/reference/random.html</loc></url>
|
||||
<url><loc>https://msberends.github.io/AMR/reference/resistance_predict.html</loc></url>
|
||||
<url><loc>https://msberends.github.io/AMR/reference/skewness.html</loc></url>
|
||||
<url><loc>https://msberends.github.io/AMR/reference/top_n_microorganisms.html</loc></url>
|
||||
<url><loc>https://msberends.github.io/AMR/reference/translate.html</loc></url>
|
||||
</urlset>
|
||||
|
||||
|
||||