# `AMR`
This is an [R package](https://www.r-project.org) to simplify the analysis and prediction of Antimicrobial Resistance (AMR).

This R package was created for academic research by PhD students of the Faculty of Medical Sciences of the [University of Groningen (rug.nl)](https://www.rug.nl/) and the Medical Microbiology & Infection Prevention department of the [University Medical Center Groningen (UMCG, umcg.nl)](https://www.umcg.nl). They also maintain this package, see [Authors](#authors).
## Why this package?
This R package contains functions to make microbiological, epidemiological data analysis easier. It allows the use of some new classes to work with MIC values and antimicrobial interpretations (i.e. values S, I and R).
With AMR you can also apply EUCAST rules to isolates, identify first isolates of every patient, translate antibiotic codes from the lab (like `"AMOX"`) or the [WHO](https://www.whocc.no/atc_ddd_index/?code=J01CA04&showdescription=no) (like `"J01CA04"`) to trivial names (like `"amoxicillin"`), or predict antimicrobial resistance for the nextcoming years with the `rsi_predict` function.
For regular AMR analysis, the `rsi` function can be used. This function als works with the `dplyr` package (e.g. in conjunction with `summarise`) to calculate the resistance percentages of different antibiotic columns of a table.
This package contains an example data set `septic_patients`, consisting of 2000 isolates from anonymised septic patients between 2001 and 2017.
## How to get it?
This package is available on CRAN and also here on GitHub.
### From CRAN (recommended)
[](http://cran.r-project.org/package=AMR)
[](http://cran.r-project.org/package=AMR)
[](http://cran.r-project.org/package=AMR)
-  In [RStudio](http://www.rstudio.com) (recommended):
- Click on `Tools` and then `Install Packages...`
- Type in `AMR` and press Install
-
In [RStudio](http://www.rstudio.com) (recommended):
- Click on `Tools` and then `Install Packages...`
- Type in `AMR` and press Install
-  In R directly:
- `install.packages("AMR")`
-
In R directly:
- `install.packages("AMR")`
-  In [Exploratory.io](https://exploratory.io):
- (Exploratory.io costs $40/month, but is free for students and teachers; if you have an `@umcg.nl` or `@rug.nl` email address, [click here to enroll](https://exploratory.io/plan?plan=Community))
- Start the software and log in
- Click on your username at the right hand side top
- Click on `R Packages`
- Click on the `Install` tab
- Type in `AMR` and press Install
- Once it’s installed it will show up in the `User Packages` section under the `Packages` tab.
### From GitHub (latest development version)
[](https://travis-ci.org/msberends/AMR)
[](https://github.com/msberends/AMR/commits/master)
[](https://github.com/msberends/AMR/commits/master)
[](https://codecov.io/gh/msberends/AMR)
```r
install.packages("devtools")
devtools::install_github("msberends/AMR")
```
## How to use it?
```r
# Call it with:
library(AMR)
# For a list of functions:
help(package = "AMR")
```
### Overwrite/force resistance based on EUCAST rules
This is also called *interpretive reading*.
```r
before <- data.frame(bactid = c("STAAUR", # Staphylococcus aureus
"ENCFAE", # Enterococcus faecalis
"ESCCOL", # Escherichia coli
"KLEPNE", # Klebsiella pneumoniae
"PSEAER"), # Pseudomonas aeruginosa
vanc = "-", # Vancomycin
amox = "-", # Amoxicillin
coli = "-", # Colistin
cfta = "-", # Ceftazidime
cfur = "-", # Cefuroxime
stringsAsFactors = FALSE)
before
# bactid vanc amox coli cfta cfur
# 1 STAAUR - - - - -
# 2 ENCFAE - - - - -
# 3 ESCCOL - - - - -
# 4 KLEPNE - - - - -
# 5 PSEAER - - - - -
# Now apply those rules; just need a column with bacteria ID's and antibiotic results:
after <- EUCAST_rules(before)
after
# bactid vanc amox coli cfta cfur
# 1 STAAUR - - R R -
# 2 ENCFAE - - R R R
# 3 ESCCOL R - - - -
# 4 KLEPNE R R - - -
# 5 PSEAER R R - - R
```
### New classes
This package contains two new S3 classes: `mic` for MIC values (e.g. from Vitek or Phoenix) and `rsi` for antimicrobial drug interpretations (i.e. S, I and R). Both are actually ordered factors under the hood (an MIC of `2` being higher than `<=1` but lower than `>=32`, and for class `rsi` factors are ordered as `S < I < R`).
Both classes have extensions for existing generic functions like `print`, `summary` and `plot`.
```r
# Transform values to new classes
mic_data <- as.mic(c(">=32", "1.0", "8", "<=0.128", "8", "16", "16"))
rsi_data <- as.rsi(c(rep("S", 474), rep("I", 36), rep("R", 370)))
```
These functions also try to coerce valid values.
Quick overviews when just printing objects:
```r
mic_data
# Class 'mic': 7 isolates
#
# 0
#
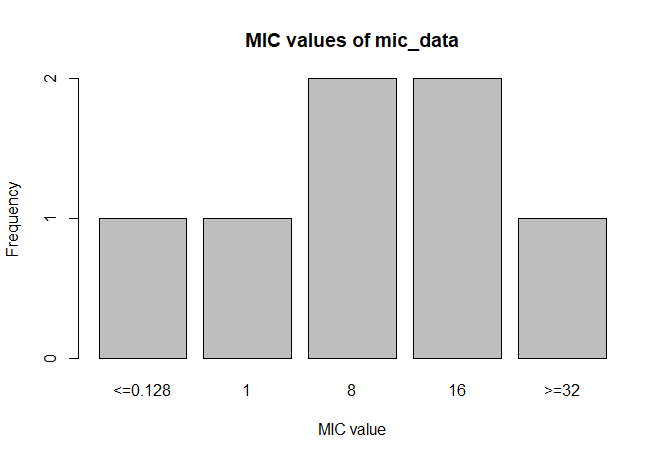
# <=0.128 1 8 16 >=32
# 1 1 2 2 1
rsi_data
# Class 'rsi': 880 isolates
#
# : 0
# Sum of S: 474
# Sum of IR: 406
# - Sum of R: 370
# - Sum of I: 36
#
# %S %IR %I %R
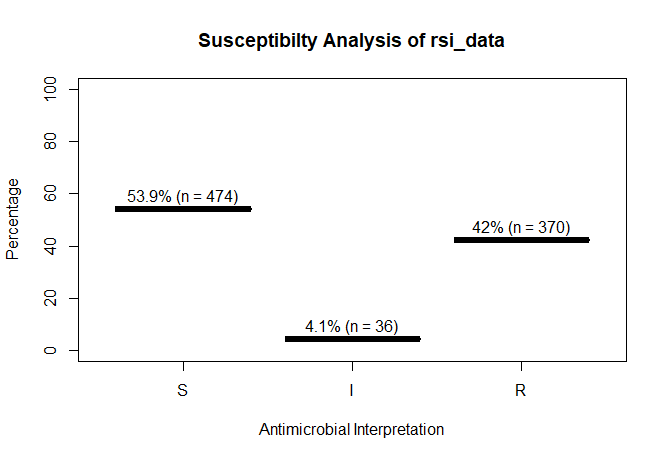
# 53.9 46.1 4.1 42.0
```
A plot of `rsi_data`:
```r
plot(rsi_data)
```
A plot of `mic_data` (defaults to bar plot):
```r
plot(mic_data)
```
Other epidemiological functions:
```r
# Determine key antibiotic based on bacteria ID
key_antibiotics(...)
# Selection of first isolates of any patient
first_isolate(...)
# Calculate resistance levels of antibiotics, can be used with `summarise` (dplyr)
rsi(...)
# Predict resistance levels of antibiotics
rsi_predict(...)
# Get name of antibiotic by ATC code
abname(...)
abname("J01CR02", from = "atc", to = "umcg") # "AMCL"
```
### Databases included in package
Datasets to work with antibiotics and bacteria properties.
```r
# Dataset with ATC antibiotics codes, official names and DDD's (oral and parenteral)
ablist # A tibble: 420 x 12
# Dataset with bacteria codes and properties like gram stain and aerobic/anaerobic
bactlist # A tibble: 2,507 x 10
```
## Authors
- [Berends MS](https://github.com/msberends)1,2, PhD Student
- [Luz CF](https://github.com/ceefluz)1, PhD Student
- [Hassing EEA](https://github.com/erwinhassing)2, Data Analyst (contributor)
1 Department of Medical Microbiology, University of Groningen, University Medical Center Groningen, Groningen, the Netherlands
2 Department of Medical, Market and Innovation (MMI), Certe Medische diagnostiek & advies, Groningen, the Netherlands
## Copyright
[](https://github.com/msberends/AMR/blob/master/LICENSE)
This R package is licensed under the [GNU General Public License (GPL) v2.0](https://github.com/msberends/AMR/blob/master/LICENSE). In a nutshell, this means that this package:
- May be used for commercial purposes
- May be used for private purposes
- May be modified, although:
- Modifications **must** be released under the same license when distributing the package
- Changes made to the code **must** be documented
- May be distributed, although:
- Source code **must** be made available when the package is distributed
- A copy of the license and copyright notice **must** be included with the package.
- Comes with a LIMITATION of liability
- Comes with NO warranty
In [Exploratory.io](https://exploratory.io):
- (Exploratory.io costs $40/month, but is free for students and teachers; if you have an `@umcg.nl` or `@rug.nl` email address, [click here to enroll](https://exploratory.io/plan?plan=Community))
- Start the software and log in
- Click on your username at the right hand side top
- Click on `R Packages`
- Click on the `Install` tab
- Type in `AMR` and press Install
- Once it’s installed it will show up in the `User Packages` section under the `Packages` tab.
### From GitHub (latest development version)
[](https://travis-ci.org/msberends/AMR)
[](https://github.com/msberends/AMR/commits/master)
[](https://github.com/msberends/AMR/commits/master)
[](https://codecov.io/gh/msberends/AMR)
```r
install.packages("devtools")
devtools::install_github("msberends/AMR")
```
## How to use it?
```r
# Call it with:
library(AMR)
# For a list of functions:
help(package = "AMR")
```
### Overwrite/force resistance based on EUCAST rules
This is also called *interpretive reading*.
```r
before <- data.frame(bactid = c("STAAUR", # Staphylococcus aureus
"ENCFAE", # Enterococcus faecalis
"ESCCOL", # Escherichia coli
"KLEPNE", # Klebsiella pneumoniae
"PSEAER"), # Pseudomonas aeruginosa
vanc = "-", # Vancomycin
amox = "-", # Amoxicillin
coli = "-", # Colistin
cfta = "-", # Ceftazidime
cfur = "-", # Cefuroxime
stringsAsFactors = FALSE)
before
# bactid vanc amox coli cfta cfur
# 1 STAAUR - - - - -
# 2 ENCFAE - - - - -
# 3 ESCCOL - - - - -
# 4 KLEPNE - - - - -
# 5 PSEAER - - - - -
# Now apply those rules; just need a column with bacteria ID's and antibiotic results:
after <- EUCAST_rules(before)
after
# bactid vanc amox coli cfta cfur
# 1 STAAUR - - R R -
# 2 ENCFAE - - R R R
# 3 ESCCOL R - - - -
# 4 KLEPNE R R - - -
# 5 PSEAER R R - - R
```
### New classes
This package contains two new S3 classes: `mic` for MIC values (e.g. from Vitek or Phoenix) and `rsi` for antimicrobial drug interpretations (i.e. S, I and R). Both are actually ordered factors under the hood (an MIC of `2` being higher than `<=1` but lower than `>=32`, and for class `rsi` factors are ordered as `S < I < R`).
Both classes have extensions for existing generic functions like `print`, `summary` and `plot`.
```r
# Transform values to new classes
mic_data <- as.mic(c(">=32", "1.0", "8", "<=0.128", "8", "16", "16"))
rsi_data <- as.rsi(c(rep("S", 474), rep("I", 36), rep("R", 370)))
```
These functions also try to coerce valid values.
Quick overviews when just printing objects:
```r
mic_data
# Class 'mic': 7 isolates
#
# 0
#
# <=0.128 1 8 16 >=32
# 1 1 2 2 1
rsi_data
# Class 'rsi': 880 isolates
#
# : 0
# Sum of S: 474
# Sum of IR: 406
# - Sum of R: 370
# - Sum of I: 36
#
# %S %IR %I %R
# 53.9 46.1 4.1 42.0
```
A plot of `rsi_data`:
```r
plot(rsi_data)
```

A plot of `mic_data` (defaults to bar plot):
```r
plot(mic_data)
```

Other epidemiological functions:
```r
# Determine key antibiotic based on bacteria ID
key_antibiotics(...)
# Selection of first isolates of any patient
first_isolate(...)
# Calculate resistance levels of antibiotics, can be used with `summarise` (dplyr)
rsi(...)
# Predict resistance levels of antibiotics
rsi_predict(...)
# Get name of antibiotic by ATC code
abname(...)
abname("J01CR02", from = "atc", to = "umcg") # "AMCL"
```
### Databases included in package
Datasets to work with antibiotics and bacteria properties.
```r
# Dataset with ATC antibiotics codes, official names and DDD's (oral and parenteral)
ablist # A tibble: 420 x 12
# Dataset with bacteria codes and properties like gram stain and aerobic/anaerobic
bactlist # A tibble: 2,507 x 10
```
## Authors
- [Berends MS](https://github.com/msberends)1,2, PhD Student
- [Luz CF](https://github.com/ceefluz)1, PhD Student
- [Hassing EEA](https://github.com/erwinhassing)2, Data Analyst (contributor)
1 Department of Medical Microbiology, University of Groningen, University Medical Center Groningen, Groningen, the Netherlands
2 Department of Medical, Market and Innovation (MMI), Certe Medische diagnostiek & advies, Groningen, the Netherlands
## Copyright
[](https://github.com/msberends/AMR/blob/master/LICENSE)
This R package is licensed under the [GNU General Public License (GPL) v2.0](https://github.com/msberends/AMR/blob/master/LICENSE). In a nutshell, this means that this package:
- May be used for commercial purposes
- May be used for private purposes
- May be modified, although:
- Modifications **must** be released under the same license when distributing the package
- Changes made to the code **must** be documented
- May be distributed, although:
- Source code **must** be made available when the package is distributed
- A copy of the license and copyright notice **must** be included with the package.
- Comes with a LIMITATION of liability
- Comes with NO warranty