Frequency table
freq.RdCreate a frequency table of a vector with items or a data frame. Supports quasiquotation and markdown for reports. The best practice is: data %>% freq(var).
top_freq can be used to get the top/bottom n items of a frequency table, with counts as names.
frequency_tbl(x, ..., sort.count = TRUE, nmax = getOption("max.print.freq"), na.rm = TRUE, row.names = TRUE, markdown = !interactive(), digits = 2, quote = FALSE, header = !markdown, title = NULL, na = "<NA>", droplevels = TRUE, sep = " ", decimal.mark = getOption("OutDec"), big.mark = ifelse(decimal.mark != ",", ",", ".")) freq(x, ..., sort.count = TRUE, nmax = getOption("max.print.freq"), na.rm = TRUE, row.names = TRUE, markdown = !interactive(), digits = 2, quote = FALSE, header = !markdown, title = NULL, na = "<NA>", droplevels = TRUE, sep = " ", decimal.mark = getOption("OutDec"), big.mark = ifelse(decimal.mark != ",", ",", ".")) top_freq(f, n) header(f, property = NULL) # S3 method for frequency_tbl print(x, nmax = getOption("max.print.freq", default = 15), markdown = !interactive(), header = !markdown, decimal.mark = getOption("OutDec"), big.mark = ifelse(decimal.mark != ",", ",", "."), ...)
Arguments
| x | vector of any class or a |
|---|---|
| ... | up to nine different columns of |
| sort.count | sort on count, i.e. frequencies. This will be |
| nmax | number of row to print. The default, |
| na.rm | a logical value indicating whether |
| row.names | a logical value indicating whether row indices should be printed as |
| markdown | a logical value indicating whether the frequency table should be printed in markdown format. This will print all rows (except when |
| digits | how many significant digits are to be used for numeric values in the header (not for the items themselves, that depends on |
| quote | a logical value indicating whether or not strings should be printed with surrounding quotes |
| header | a logical value indicating whether an informative header should be printed |
| title | text to show above frequency table, at default to tries to coerce from the variables passed to |
| na | a character string that should be used to show empty ( |
| droplevels | a logical value indicating whether in factors empty levels should be dropped |
| sep | a character string to separate the terms when selecting multiple columns |
| decimal.mark |
used for prettying (longish) numerical and complex sequences.
Passed to |
| big.mark |
used for prettying (longish) numerical and complex sequences.
Passed to |
| f | a frequency table |
| n | number of top n items to return, use -n for the bottom n items. It will include more than |
| property | property in header to return this value directly |
Value
A data.frame (with an additional class "frequency_tbl") with five columns: item, count, percent, cum_count and cum_percent.
Details
Frequency tables (or frequency distributions) are summaries of the distribution of values in a sample. With the `freq` function, you can create univariate frequency tables. Multiple variables will be pasted into one variable, so it forces a univariate distribution. This package also has a vignette available to explain the use of this function further, run browseVignettes("AMR") to read it.
For numeric values of any class, these additional values will all be calculated with na.rm = TRUE and shown into the header:
Mean, using
meanStandard Deviation, using
sdCoefficient of Variation (CV), the standard deviation divided by the mean
Mean Absolute Deviation (MAD), using
madTukey Five-Number Summaries (minimum, Q1, median, Q3, maximum), using
fivenumInterquartile Range (IQR) calculated as
Q3 - Q1using the Tukey Five-Number Summaries, i.e. not using thequantilefunctionCoefficient of Quartile Variation (CQV, sometimes called coefficient of dispersion), calculated as
(Q3 - Q1) / (Q3 + Q1)using the Tukey Five-Number SummariesOutliers (total count and unique count), using
boxplot.stats
For dates and times of any class, these additional values will be calculated with na.rm = TRUE and shown into the header:
Oldest, using
minNewest, using
max, with difference between newest and oldestMedian, using
median, with percentage since oldest
In factors, all factor levels that are not existing in the input data will be dropped.
The function top_freq uses top_n internally and will include more than n rows if there are ties.
Examples
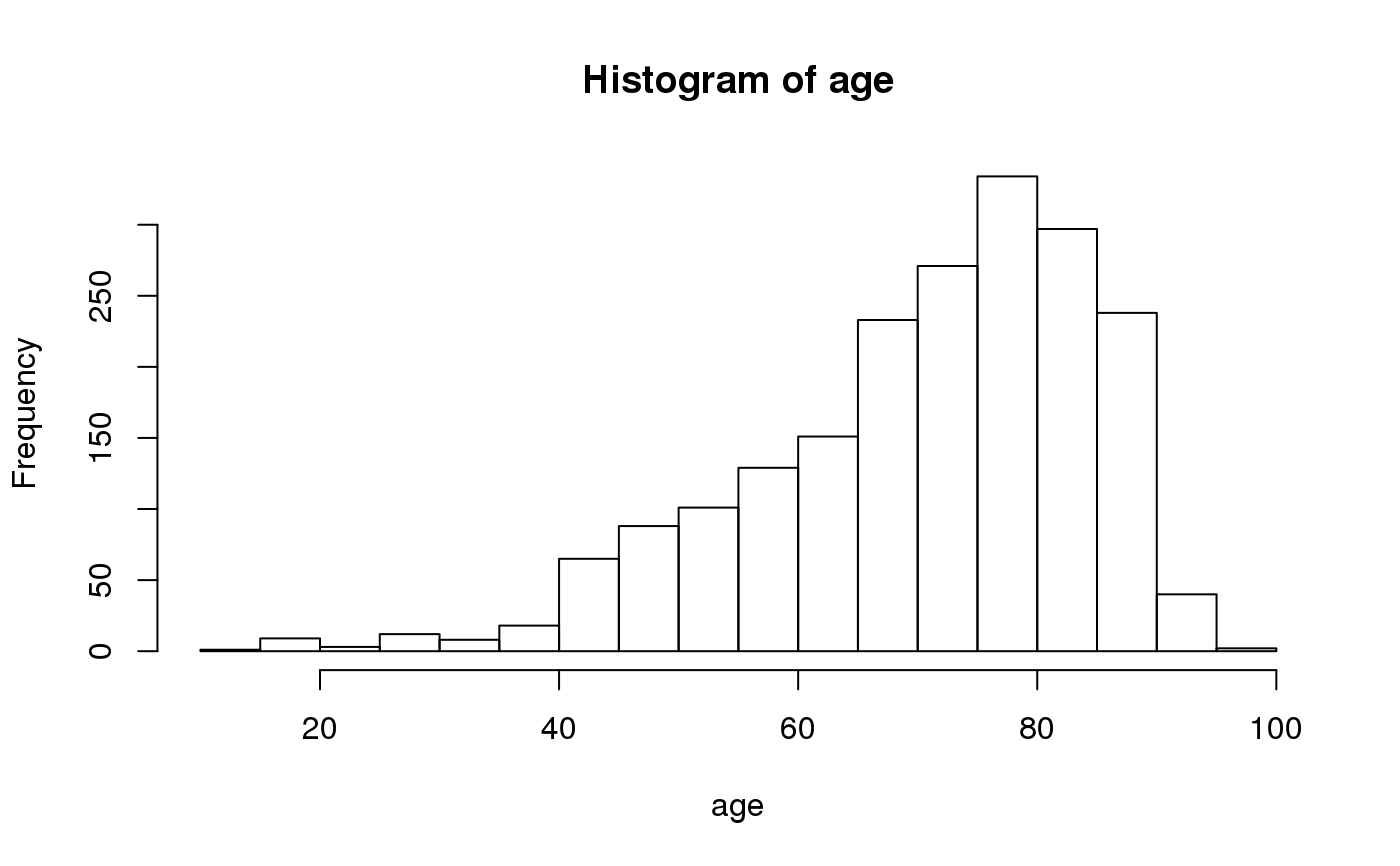
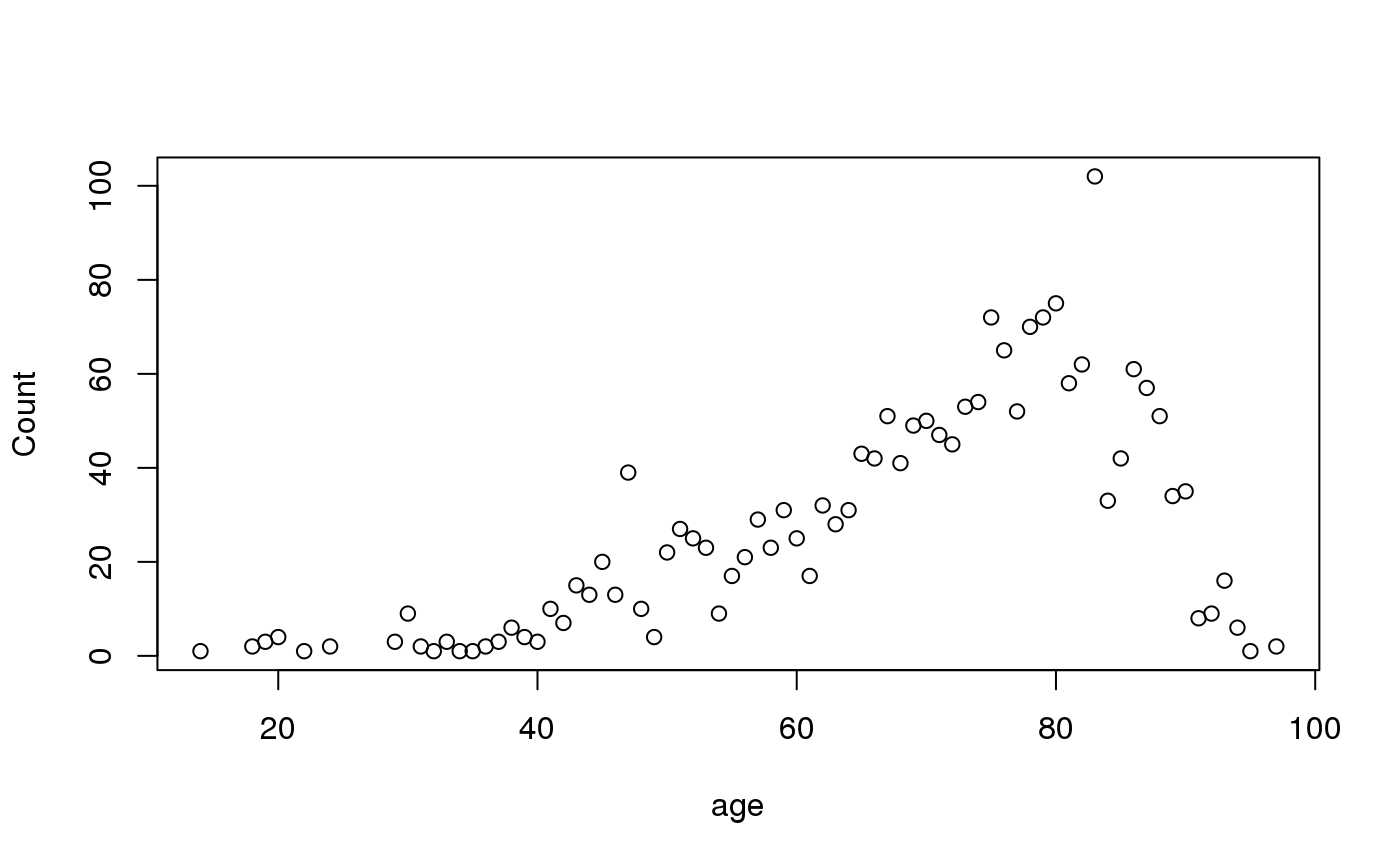
#> #> #> **Frequency table** #> #> #> | |Item | Count| Percent| Cum. Count| Cum. Percent| #> |:--|:----|-----:|-------:|----------:|------------:| #> |1 |D | 762| 38.1%| 762| 38.1%| #> |2 |B | 663| 33.2%| 1,425| 71.3%| #> |3 |A | 321| 16.1%| 1,746| 87.3%| #> |4 |C | 254| 12.7%| 2,000| 100.0%| #> #>freq(septic_patients[, "hospital_id"])#> #> #> **Frequency table** #> #> #> | |Item | Count| Percent| Cum. Count| Cum. Percent| #> |:--|:----|-----:|-------:|----------:|------------:| #> |1 |D | 762| 38.1%| 762| 38.1%| #> |2 |B | 663| 33.2%| 1,425| 71.3%| #> |3 |A | 321| 16.1%| 1,746| 87.3%| #> |4 |C | 254| 12.7%| 2,000| 100.0%| #> #>septic_patients$hospital_id %>% freq()#> #> #> **Frequency table** #> #> #> | |Item | Count| Percent| Cum. Count| Cum. Percent| #> |:--|:----|-----:|-------:|----------:|------------:| #> |1 |D | 762| 38.1%| 762| 38.1%| #> |2 |B | 663| 33.2%| 1,425| 71.3%| #> |3 |A | 321| 16.1%| 1,746| 87.3%| #> |4 |C | 254| 12.7%| 2,000| 100.0%| #> #>septic_patients[, "hospital_id"] %>% freq()#> #> #> **Frequency table** #> #> #> | |Item | Count| Percent| Cum. Count| Cum. Percent| #> |:--|:----|-----:|-------:|----------:|------------:| #> |1 |D | 762| 38.1%| 762| 38.1%| #> |2 |B | 663| 33.2%| 1,425| 71.3%| #> |3 |A | 321| 16.1%| 1,746| 87.3%| #> |4 |C | 254| 12.7%| 2,000| 100.0%| #> #>septic_patients %>% freq("hospital_id")#> #> #> **Frequency table of `hospital_id`** #> #> #> | |Item | Count| Percent| Cum. Count| Cum. Percent| #> |:--|:----|-----:|-------:|----------:|------------:| #> |1 |D | 762| 38.1%| 762| 38.1%| #> |2 |B | 663| 33.2%| 1,425| 71.3%| #> |3 |A | 321| 16.1%| 1,746| 87.3%| #> |4 |C | 254| 12.7%| 2,000| 100.0%| #> #>septic_patients %>% freq(hospital_id) #<- easiest to remember (tidyverse)#> #> #> **Frequency table of `hospital_id`** #> #> #> | |Item | Count| Percent| Cum. Count| Cum. Percent| #> |:--|:----|-----:|-------:|----------:|------------:| #> |1 |D | 762| 38.1%| 762| 38.1%| #> |2 |B | 663| 33.2%| 1,425| 71.3%| #> |3 |A | 321| 16.1%| 1,746| 87.3%| #> |4 |C | 254| 12.7%| 2,000| 100.0%| #> #># you could also use `select` or `pull` to get your variables septic_patients %>% filter(hospital_id == "A") %>% select(mo) %>% freq()#> #> #> **Frequency table** #> #> #> | |Item | Count| Percent| Cum. Count| Cum. Percent| #> |:--|:------------|-----:|-------:|----------:|------------:| #> |1 |B_ESCHR_COL | 62| 19.3%| 62| 19.3%| #> |2 |B_STPHY_EPI | 46| 14.3%| 108| 33.6%| #> |3 |B_STPHY_CNS | 38| 11.8%| 146| 45.5%| #> |4 |B_STPHY_AUR | 35| 10.9%| 181| 56.4%| #> |5 |B_STPHY_HOM | 25| 7.8%| 206| 64.2%| #> |6 |B_STRPTC_PNE | 12| 3.7%| 218| 67.9%| #> |7 |B_PROTS_MIR | 11| 3.4%| 229| 71.3%| #> |8 |B_ENTRC_FAE | 10| 3.1%| 239| 74.5%| #> |9 |B_KLBSL_PNE | 8| 2.5%| 247| 76.9%| #> |10 |B_STRPTC_PYO | 7| 2.2%| 254| 79.1%| #> |11 |B_BCTRD_FRA | 5| 1.6%| 259| 80.7%| #> |12 |B_KLBSL_OXY | 5| 1.6%| 264| 82.2%| #> |13 |B_STRPTC | 5| 1.6%| 269| 83.8%| #> |14 |B_ENTRC_IUM | 4| 1.2%| 273| 85.0%| #> |15 |B_STRPTC_MIT | 4| 1.2%| 277| 86.3%| #> |16 |B_CRYNB | 3| 0.9%| 280| 87.2%| #> |17 |B_PDMNS_AER | 3| 0.9%| 283| 88.2%| #> |18 |B_STPHY_CAP | 3| 0.9%| 286| 89.1%| #> |19 |B_STRPTC_DYS | 3| 0.9%| 289| 90.0%| #> |20 |F_CANDD_GLB | 3| 0.9%| 292| 91.0%| #> |21 |B_ACNTB | 2| 0.6%| 294| 91.6%| #> |22 |B_ENTRB_CLO | 2| 0.6%| 296| 92.2%| #> |23 |B_HMPHL_INF | 2| 0.6%| 298| 92.8%| #> |24 |B_MCRCCC | 2| 0.6%| 300| 93.5%| #> |25 |B_PROTS_VUL | 2| 0.6%| 302| 94.1%| #> |26 |B_SERRT_MAR | 2| 0.6%| 304| 94.7%| #> |27 |B_STPHY_COH | 2| 0.6%| 306| 95.3%| #> |28 |B_STRPTC_BOV | 2| 0.6%| 308| 96.0%| #> |29 |B_AMYCS_ODO | 1| 0.3%| 309| 96.3%| #> |30 |B_ARCCC_URI | 1| 0.3%| 310| 96.6%| #> |31 |B_CTRDM_PER | 1| 0.3%| 311| 96.9%| #> |32 |B_CTRDM_SEP | 1| 0.3%| 312| 97.2%| #> |33 |B_STPHY_SCH | 1| 0.3%| 313| 97.5%| #> |34 |B_STRPTC_AGA | 1| 0.3%| 314| 97.8%| #> |35 |B_STRPTC_EQU | 1| 0.3%| 315| 98.1%| #> |36 |B_STRPTC_GRA | 1| 0.3%| 316| 98.4%| #> |37 |B_STRPTC_GRB | 1| 0.3%| 317| 98.8%| #> |38 |B_STRPTC_SAN | 1| 0.3%| 318| 99.1%| #> |39 |B_VLLNL_PAR | 1| 0.3%| 319| 99.4%| #> |40 |F_CANDD_ALB | 1| 0.3%| 320| 99.7%| #> |41 |F_CANDD_TRO | 1| 0.3%| 321| 100.0%| #> #># multiple selected variables will be pasted together septic_patients %>% left_join_microorganisms %>% filter(hospital_id == "A") %>% freq(genus, species)#>#> #> #> **Frequency table of `genus` and `species`** #> #> #> | |Item | Count| Percent| Cum. Count| Cum. Percent| #> |:--|:---------------------------------|-----:|-------:|----------:|------------:| #> |1 |Escherichia coli | 62| 19.3%| 62| 19.3%| #> |2 |Staphylococcus epidermidis | 46| 14.3%| 108| 33.6%| #> |3 |Staphylococcus coagulase negative | 38| 11.8%| 146| 45.5%| #> |4 |Staphylococcus aureus | 35| 10.9%| 181| 56.4%| #> |5 |Staphylococcus hominis | 25| 7.8%| 206| 64.2%| #> |6 |Streptococcus pneumoniae | 12| 3.7%| 218| 67.9%| #> |7 |Proteus mirabilis | 11| 3.4%| 229| 71.3%| #> |8 |Enterococcus faecalis | 10| 3.1%| 239| 74.5%| #> |9 |Klebsiella pneumoniae | 8| 2.5%| 247| 76.9%| #> |10 |Streptococcus pyogenes | 7| 2.2%| 254| 79.1%| #> |11 |Bacteroides fragilis | 5| 1.6%| 259| 80.7%| #> |12 |Klebsiella oxytoca | 5| 1.6%| 264| 82.2%| #> |13 |Streptococcus species | 5| 1.6%| 269| 83.8%| #> |14 |Enterococcus faecium | 4| 1.2%| 273| 85.0%| #> |15 |Streptococcus mitis | 4| 1.2%| 277| 86.3%| #> |16 |Candida glabrata | 3| 0.9%| 280| 87.2%| #> |17 |Corynebacterium species | 3| 0.9%| 283| 88.2%| #> |18 |Pseudomonas aeruginosa | 3| 0.9%| 286| 89.1%| #> |19 |Staphylococcus capitis | 3| 0.9%| 289| 90.0%| #> |20 |Streptococcus dysgalactiae | 3| 0.9%| 292| 91.0%| #> |21 |Acinetobacter species | 2| 0.6%| 294| 91.6%| #> |22 |Enterobacter cloacae | 2| 0.6%| 296| 92.2%| #> |23 |Haemophilus influenzae | 2| 0.6%| 298| 92.8%| #> |24 |Micrococcus species | 2| 0.6%| 300| 93.5%| #> |25 |Proteus vulgaris | 2| 0.6%| 302| 94.1%| #> |26 |Serratia marcescens | 2| 0.6%| 304| 94.7%| #> |27 |Staphylococcus cohnii | 2| 0.6%| 306| 95.3%| #> |28 |Streptococcus bovis | 2| 0.6%| 308| 96.0%| #> |29 |Actinomyces odontolyticus | 1| 0.3%| 309| 96.3%| #> |30 |Aerococcus urinae | 1| 0.3%| 310| 96.6%| #> |31 |Candida albicans | 1| 0.3%| 311| 96.9%| #> |32 |Candida tropicalis | 1| 0.3%| 312| 97.2%| #> |33 |Clostridium perfringens | 1| 0.3%| 313| 97.5%| #> |34 |Clostridium septicum | 1| 0.3%| 314| 97.8%| #> |35 |Staphylococcus schleiferi | 1| 0.3%| 315| 98.1%| #> |36 |Streptococcus agalactiae | 1| 0.3%| 316| 98.4%| #> |37 |Streptococcus equi | 1| 0.3%| 317| 98.8%| #> |38 |Streptococcus group A | 1| 0.3%| 318| 99.1%| #> |39 |Streptococcus group B | 1| 0.3%| 319| 99.4%| #> |40 |Streptococcus sanguinis | 1| 0.3%| 320| 99.7%| #> |41 |Veillonella parvula | 1| 0.3%| 321| 100.0%| #> #>#> #> #> **Frequency table of `gender` (grouped by `hospital_id`)** #> #> #> | |Group |Item | Count| Percent| Cum. Count| Cum. Percent| #> |:--|:-----|:----|-----:|-------:|----------:|------------:| #> |1 |1 |F | 148| 7.4%| 148| 7.4%| #> |2 | |M | 173| 8.7%| 321| 16.1%| #> |3 |2 |F | 332| 16.6%| 332| 16.6%| #> |4 | |M | 331| 16.6%| 663| 33.2%| #> |5 |3 |F | 121| 6.1%| 121| 6.1%| #> |6 | |M | 133| 6.7%| 254| 12.7%| #> |7 |4 |F | 368| 18.4%| 368| 18.4%| #> |8 | |M | 394| 19.7%| 762| 38.1%| #> #># get top 10 bugs of hospital A as a vector septic_patients %>% filter(hospital_id == "A") %>% freq(mo) %>% top_freq(10)#> 62 46 38 35 25 #> "B_ESCHR_COL" "B_STPHY_EPI" "B_STPHY_CNS" "B_STPHY_AUR" "B_STPHY_HOM" #> 12 11 10 8 7 #> "B_STRPTC_PNE" "B_PROTS_MIR" "B_ENTRC_FAE" "B_KLBSL_PNE" "B_STRPTC_PYO"# save frequency table to an object years <- septic_patients %>% mutate(year = format(date, "%Y")) %>% freq(year) # show only the top 5 years %>% print(nmax = 5)#> #> #> **Frequency table of `year`** #> #> #> | |Item | Count| Percent| Cum. Count| Cum. Percent| #> |:--|:----|-----:|-------:|----------:|------------:| #> |1 |2017 | 168| 8.4%| 168| 8.4%| #> |2 |2004 | 167| 8.4%| 335| 16.8%| #> |3 |2016 | 143| 7.2%| 478| 23.9%| #> |4 |2002 | 136| 6.8%| 614| 30.7%| #> |5 |2003 | 135| 6.8%| 749| 37.5%| #> #> (omitted 11 entries, n = 1,251 [62.6%]) #> #># save to an object with formatted percentages years <- format(years) # print a histogram of numeric values septic_patients %>% freq(age) %>% hist()#> item count percent cum_count cum_percent #> 1 83 102 0.0510 102 0.0510 #> 2 80 75 0.0375 177 0.0885 #> 3 75 72 0.0360 249 0.1245 #> 4 79 72 0.0360 321 0.1605 #> 5 78 70 0.0350 391 0.1955 #> 6 76 65 0.0325 456 0.2280 #> 7 82 62 0.0310 518 0.2590 #> 8 86 61 0.0305 579 0.2895 #> 9 81 58 0.0290 637 0.3185 #> 10 87 57 0.0285 694 0.3470 #> 11 74 54 0.0270 748 0.3740 #> 12 73 53 0.0265 801 0.4005 #> 13 77 52 0.0260 853 0.4265 #> 14 67 51 0.0255 904 0.4520 #> 15 88 51 0.0255 955 0.4775 #> 16 70 50 0.0250 1005 0.5025 #> 17 69 49 0.0245 1054 0.5270 #> 18 71 47 0.0235 1101 0.5505 #> 19 72 45 0.0225 1146 0.5730 #> 20 65 43 0.0215 1189 0.5945 #> 21 66 42 0.0210 1231 0.6155 #> 22 85 42 0.0210 1273 0.6365 #> 23 68 41 0.0205 1314 0.6570 #> 24 47 39 0.0195 1353 0.6765 #> 25 90 35 0.0175 1388 0.6940 #> 26 89 34 0.0170 1422 0.7110 #> 27 84 33 0.0165 1455 0.7275 #> 28 62 32 0.0160 1487 0.7435 #> 29 59 31 0.0155 1518 0.7590 #> 30 64 31 0.0155 1549 0.7745 #> 31 57 29 0.0145 1578 0.7890 #> 32 63 28 0.0140 1606 0.8030 #> 33 51 27 0.0135 1633 0.8165 #> 34 52 25 0.0125 1658 0.8290 #> 35 60 25 0.0125 1683 0.8415 #> 36 53 23 0.0115 1706 0.8530 #> 37 58 23 0.0115 1729 0.8645 #> 38 50 22 0.0110 1751 0.8755 #> 39 56 21 0.0105 1772 0.8860 #> 40 45 20 0.0100 1792 0.8960 #> 41 55 17 0.0085 1809 0.9045 #> 42 61 17 0.0085 1826 0.9130 #> 43 93 16 0.0080 1842 0.9210 #> 44 43 15 0.0075 1857 0.9285 #> 45 44 13 0.0065 1870 0.9350 #> 46 46 13 0.0065 1883 0.9415 #> 47 41 10 0.0050 1893 0.9465 #> 48 48 10 0.0050 1903 0.9515 #> 49 30 9 0.0045 1912 0.9560 #> 50 54 9 0.0045 1921 0.9605 #> 51 92 9 0.0045 1930 0.9650 #> 52 91 8 0.0040 1938 0.9690 #> 53 42 7 0.0035 1945 0.9725 #> 54 38 6 0.0030 1951 0.9755 #> 55 94 6 0.0030 1957 0.9785 #> 56 20 4 0.0020 1961 0.9805 #> 57 39 4 0.0020 1965 0.9825 #> 58 49 4 0.0020 1969 0.9845 #> 59 19 3 0.0015 1972 0.9860 #> 60 29 3 0.0015 1975 0.9875 #> 61 33 3 0.0015 1978 0.9890 #> 62 37 3 0.0015 1981 0.9905 #> 63 40 3 0.0015 1984 0.9920 #> 64 18 2 0.0010 1986 0.9930 #> 65 24 2 0.0010 1988 0.9940 #> 66 31 2 0.0010 1990 0.9950 #> 67 36 2 0.0010 1992 0.9960 #> 68 97 2 0.0010 1994 0.9970 #> 69 14 1 0.0005 1995 0.9975 #> 70 22 1 0.0005 1996 0.9980 #> 71 32 1 0.0005 1997 0.9985 #> 72 34 1 0.0005 1998 0.9990 #> 73 35 1 0.0005 1999 0.9995 #> 74 95 1 0.0005 2000 1.0000#> [1] 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 #> [25] 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 #> [49] 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 #> [73] 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 83 #> [97] 83 83 83 83 83 83 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 #> [121] 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 #> [145] 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 #> [169] 80 80 80 80 80 80 80 80 80 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 #> [193] 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 #> [217] 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 75 #> [241] 75 75 75 75 75 75 75 75 75 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 #> [265] 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 #> [289] 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 79 #> [313] 79 79 79 79 79 79 79 79 79 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 #> [337] 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 #> [361] 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 78 #> [385] 78 78 78 78 78 78 78 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 #> [409] 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 #> [433] 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 76 #> [457] 82 82 82 82 82 82 82 82 82 82 82 82 82 82 82 82 82 82 82 82 82 82 82 82 #> [481] 82 82 82 82 82 82 82 82 82 82 82 82 82 82 82 82 82 82 82 82 82 82 82 82 #> [505] 82 82 82 82 82 82 82 82 82 82 82 82 82 82 86 86 86 86 86 86 86 86 86 86 #> [529] 86 86 86 86 86 86 86 86 86 86 86 86 86 86 86 86 86 86 86 86 86 86 86 86 #> [553] 86 86 86 86 86 86 86 86 86 86 86 86 86 86 86 86 86 86 86 86 86 86 86 86 #> [577] 86 86 86 81 81 81 81 81 81 81 81 81 81 81 81 81 81 81 81 81 81 81 81 81 #> [601] 81 81 81 81 81 81 81 81 81 81 81 81 81 81 81 81 81 81 81 81 81 81 81 81 #> [625] 81 81 81 81 81 81 81 81 81 81 81 81 81 87 87 87 87 87 87 87 87 87 87 87 #> [649] 87 87 87 87 87 87 87 87 87 87 87 87 87 87 87 87 87 87 87 87 87 87 87 87 #> [673] 87 87 87 87 87 87 87 87 87 87 87 87 87 87 87 87 87 87 87 87 87 87 74 74 #> [697] 74 74 74 74 74 74 74 74 74 74 74 74 74 74 74 74 74 74 74 74 74 74 74 74 #> [721] 74 74 74 74 74 74 74 74 74 74 74 74 74 74 74 74 74 74 74 74 74 74 74 74 #> [745] 74 74 74 74 73 73 73 73 73 73 73 73 73 73 73 73 73 73 73 73 73 73 73 73 #> [769] 73 73 73 73 73 73 73 73 73 73 73 73 73 73 73 73 73 73 73 73 73 73 73 73 #> [793] 73 73 73 73 73 73 73 73 73 77 77 77 77 77 77 77 77 77 77 77 77 77 77 77 #> [817] 77 77 77 77 77 77 77 77 77 77 77 77 77 77 77 77 77 77 77 77 77 77 77 77 #> [841] 77 77 77 77 77 77 77 77 77 77 77 77 77 67 67 67 67 67 67 67 67 67 67 67 #> [865] 67 67 67 67 67 67 67 67 67 67 67 67 67 67 67 67 67 67 67 67 67 67 67 67 #> [889] 67 67 67 67 67 67 67 67 67 67 67 67 67 67 67 67 88 88 88 88 88 88 88 88 #> [913] 88 88 88 88 88 88 88 88 88 88 88 88 88 88 88 88 88 88 88 88 88 88 88 88 #> [937] 88 88 88 88 88 88 88 88 88 88 88 88 88 88 88 88 88 88 88 70 70 70 70 70 #> [961] 70 70 70 70 70 70 70 70 70 70 70 70 70 70 70 70 70 70 70 70 70 70 70 70 #> [985] 70 70 70 70 70 70 70 70 70 70 70 70 70 70 70 70 70 70 70 70 70 69 69 69 #> [1009] 69 69 69 69 69 69 69 69 69 69 69 69 69 69 69 69 69 69 69 69 69 69 69 69 #> [1033] 69 69 69 69 69 69 69 69 69 69 69 69 69 69 69 69 69 69 69 69 69 69 71 71 #> [1057] 71 71 71 71 71 71 71 71 71 71 71 71 71 71 71 71 71 71 71 71 71 71 71 71 #> [1081] 71 71 71 71 71 71 71 71 71 71 71 71 71 71 71 71 71 71 71 71 71 72 72 72 #> [1105] 72 72 72 72 72 72 72 72 72 72 72 72 72 72 72 72 72 72 72 72 72 72 72 72 #> [1129] 72 72 72 72 72 72 72 72 72 72 72 72 72 72 72 72 72 72 65 65 65 65 65 65 #> [1153] 65 65 65 65 65 65 65 65 65 65 65 65 65 65 65 65 65 65 65 65 65 65 65 65 #> [1177] 65 65 65 65 65 65 65 65 65 65 65 65 65 66 66 66 66 66 66 66 66 66 66 66 #> [1201] 66 66 66 66 66 66 66 66 66 66 66 66 66 66 66 66 66 66 66 66 66 66 66 66 #> [1225] 66 66 66 66 66 66 66 85 85 85 85 85 85 85 85 85 85 85 85 85 85 85 85 85 #> [1249] 85 85 85 85 85 85 85 85 85 85 85 85 85 85 85 85 85 85 85 85 85 85 85 85 #> [1273] 85 68 68 68 68 68 68 68 68 68 68 68 68 68 68 68 68 68 68 68 68 68 68 68 #> [1297] 68 68 68 68 68 68 68 68 68 68 68 68 68 68 68 68 68 68 47 47 47 47 47 47 #> [1321] 47 47 47 47 47 47 47 47 47 47 47 47 47 47 47 47 47 47 47 47 47 47 47 47 #> [1345] 47 47 47 47 47 47 47 47 47 90 90 90 90 90 90 90 90 90 90 90 90 90 90 90 #> [1369] 90 90 90 90 90 90 90 90 90 90 90 90 90 90 90 90 90 90 90 90 89 89 89 89 #> [1393] 89 89 89 89 89 89 89 89 89 89 89 89 89 89 89 89 89 89 89 89 89 89 89 89 #> [1417] 89 89 89 89 89 89 84 84 84 84 84 84 84 84 84 84 84 84 84 84 84 84 84 84 #> [1441] 84 84 84 84 84 84 84 84 84 84 84 84 84 84 84 62 62 62 62 62 62 62 62 62 #> [1465] 62 62 62 62 62 62 62 62 62 62 62 62 62 62 62 62 62 62 62 62 62 62 62 59 #> [1489] 59 59 59 59 59 59 59 59 59 59 59 59 59 59 59 59 59 59 59 59 59 59 59 59 #> [1513] 59 59 59 59 59 59 64 64 64 64 64 64 64 64 64 64 64 64 64 64 64 64 64 64 #> [1537] 64 64 64 64 64 64 64 64 64 64 64 64 64 57 57 57 57 57 57 57 57 57 57 57 #> [1561] 57 57 57 57 57 57 57 57 57 57 57 57 57 57 57 57 57 57 63 63 63 63 63 63 #> [1585] 63 63 63 63 63 63 63 63 63 63 63 63 63 63 63 63 63 63 63 63 63 63 51 51 #> [1609] 51 51 51 51 51 51 51 51 51 51 51 51 51 51 51 51 51 51 51 51 51 51 51 51 #> [1633] 51 52 52 52 52 52 52 52 52 52 52 52 52 52 52 52 52 52 52 52 52 52 52 52 #> [1657] 52 52 60 60 60 60 60 60 60 60 60 60 60 60 60 60 60 60 60 60 60 60 60 60 #> [1681] 60 60 60 53 53 53 53 53 53 53 53 53 53 53 53 53 53 53 53 53 53 53 53 53 #> [1705] 53 53 58 58 58 58 58 58 58 58 58 58 58 58 58 58 58 58 58 58 58 58 58 58 #> [1729] 58 50 50 50 50 50 50 50 50 50 50 50 50 50 50 50 50 50 50 50 50 50 50 56 #> [1753] 56 56 56 56 56 56 56 56 56 56 56 56 56 56 56 56 56 56 56 56 45 45 45 45 #> [1777] 45 45 45 45 45 45 45 45 45 45 45 45 45 45 45 45 55 55 55 55 55 55 55 55 #> [1801] 55 55 55 55 55 55 55 55 55 61 61 61 61 61 61 61 61 61 61 61 61 61 61 61 #> [1825] 61 61 93 93 93 93 93 93 93 93 93 93 93 93 93 93 93 93 43 43 43 43 43 43 #> [1849] 43 43 43 43 43 43 43 43 43 44 44 44 44 44 44 44 44 44 44 44 44 44 46 46 #> [1873] 46 46 46 46 46 46 46 46 46 46 46 41 41 41 41 41 41 41 41 41 41 48 48 48 #> [1897] 48 48 48 48 48 48 48 30 30 30 30 30 30 30 30 30 54 54 54 54 54 54 54 54 #> [1921] 54 92 92 92 92 92 92 92 92 92 91 91 91 91 91 91 91 91 42 42 42 42 42 42 #> [1945] 42 38 38 38 38 38 38 94 94 94 94 94 94 20 20 20 20 39 39 39 39 49 49 49 #> [1969] 49 19 19 19 29 29 29 33 33 33 37 37 37 40 40 40 18 18 24 24 31 31 36 36 #> [1993] 97 97 14 22 32 34 35 95identical(septic_patients %>% freq(age) %>% as.vector() %>% sort(), sort(septic_patients$age)) # TRUE#> [1] TRUE# it also supports `table` objects table(septic_patients$gender, septic_patients$age) %>% freq(sep = " **sep** ")#> #> #> **Frequency table of a `table` object** #> #> #> | |Item | Count| Percent| Cum. Count| Cum. Percent| #> |:---|:------------|-----:|-------:|----------:|------------:| #> |1 |F **sep** 83 | 55| 2.8%| 55| 2.8%| #> |2 |M **sep** 78 | 51| 2.6%| 106| 5.3%| #> |3 |M **sep** 83 | 47| 2.4%| 153| 7.7%| #> |4 |M **sep** 82 | 43| 2.2%| 196| 9.8%| #> |5 |M **sep** 79 | 42| 2.1%| 238| 11.9%| #> |6 |F **sep** 80 | 41| 2.1%| 279| 14.0%| #> |7 |F **sep** 76 | 39| 2.0%| 318| 15.9%| #> |8 |F **sep** 75 | 38| 1.9%| 356| 17.8%| #> |9 |F **sep** 86 | 38| 1.9%| 394| 19.7%| #> |10 |M **sep** 71 | 35| 1.8%| 429| 21.5%| #> |11 |M **sep** 75 | 34| 1.7%| 463| 23.2%| #> |12 |M **sep** 77 | 34| 1.7%| 497| 24.9%| #> |13 |M **sep** 80 | 34| 1.7%| 531| 26.6%| #> |14 |F **sep** 81 | 33| 1.7%| 564| 28.2%| #> |15 |M **sep** 88 | 31| 1.6%| 595| 29.8%| #> |16 |F **sep** 79 | 30| 1.5%| 625| 31.3%| #> |17 |M **sep** 74 | 30| 1.5%| 655| 32.8%| #> |18 |M **sep** 87 | 30| 1.5%| 685| 34.3%| #> |19 |F **sep** 65 | 29| 1.5%| 714| 35.7%| #> |20 |M **sep** 73 | 29| 1.5%| 743| 37.2%| #> |21 |M **sep** 69 | 28| 1.4%| 771| 38.6%| #> |22 |M **sep** 72 | 28| 1.4%| 799| 40.0%| #> |23 |F **sep** 67 | 27| 1.4%| 826| 41.3%| #> |24 |F **sep** 87 | 27| 1.4%| 853| 42.7%| #> |25 |M **sep** 76 | 26| 1.3%| 879| 44.0%| #> |26 |F **sep** 66 | 25| 1.3%| 904| 45.2%| #> |27 |F **sep** 70 | 25| 1.3%| 929| 46.5%| #> |28 |M **sep** 70 | 25| 1.3%| 954| 47.7%| #> |29 |M **sep** 81 | 25| 1.3%| 979| 49.0%| #> |30 |F **sep** 47 | 24| 1.2%| 1,003| 50.1%| #> |31 |F **sep** 73 | 24| 1.2%| 1,027| 51.4%| #> |32 |F **sep** 74 | 24| 1.2%| 1,051| 52.6%| #> |33 |M **sep** 67 | 24| 1.2%| 1,075| 53.8%| #> |34 |M **sep** 86 | 23| 1.2%| 1,098| 54.9%| #> |35 |F **sep** 89 | 22| 1.1%| 1,120| 56.0%| #> |36 |M **sep** 68 | 22| 1.1%| 1,142| 57.1%| #> |37 |M **sep** 85 | 22| 1.1%| 1,164| 58.2%| #> |38 |F **sep** 69 | 21| 1.1%| 1,185| 59.3%| #> |39 |M **sep** 62 | 21| 1.1%| 1,206| 60.3%| #> |40 |F **sep** 85 | 20| 1.0%| 1,226| 61.3%| #> |41 |F **sep** 88 | 20| 1.0%| 1,246| 62.3%| #> |42 |F **sep** 90 | 20| 1.0%| 1,266| 63.3%| #> |43 |M **sep** 64 | 20| 1.0%| 1,286| 64.3%| #> |44 |F **sep** 68 | 19| 1.0%| 1,305| 65.3%| #> |45 |F **sep** 78 | 19| 1.0%| 1,324| 66.2%| #> |46 |F **sep** 82 | 19| 1.0%| 1,343| 67.2%| #> |47 |F **sep** 45 | 18| 0.9%| 1,361| 68.1%| #> |48 |F **sep** 60 | 18| 0.9%| 1,379| 69.0%| #> |49 |F **sep** 77 | 18| 0.9%| 1,397| 69.9%| #> |50 |F **sep** 84 | 18| 0.9%| 1,415| 70.8%| #> |51 |M **sep** 51 | 18| 0.9%| 1,433| 71.7%| #> |52 |M **sep** 52 | 18| 0.9%| 1,451| 72.6%| #> |53 |F **sep** 57 | 17| 0.9%| 1,468| 73.4%| #> |54 |F **sep** 72 | 17| 0.9%| 1,485| 74.3%| #> |55 |M **sep** 66 | 17| 0.9%| 1,502| 75.1%| #> |56 |F **sep** 58 | 16| 0.8%| 1,518| 75.9%| #> |57 |F **sep** 59 | 16| 0.8%| 1,534| 76.7%| #> |58 |M **sep** 56 | 16| 0.8%| 1,550| 77.5%| #> |59 |M **sep** 47 | 15| 0.8%| 1,565| 78.3%| #> |60 |M **sep** 59 | 15| 0.8%| 1,580| 79.0%| #> |61 |M **sep** 84 | 15| 0.8%| 1,595| 79.8%| #> |62 |M **sep** 90 | 15| 0.8%| 1,610| 80.5%| #> |63 |F **sep** 63 | 14| 0.7%| 1,624| 81.2%| #> |64 |M **sep** 53 | 14| 0.7%| 1,638| 81.9%| #> |65 |M **sep** 63 | 14| 0.7%| 1,652| 82.6%| #> |66 |M **sep** 65 | 14| 0.7%| 1,666| 83.3%| #> |67 |M **sep** 61 | 13| 0.7%| 1,679| 84.0%| #> |68 |F **sep** 50 | 12| 0.6%| 1,691| 84.6%| #> |69 |F **sep** 71 | 12| 0.6%| 1,703| 85.2%| #> |70 |M **sep** 57 | 12| 0.6%| 1,715| 85.8%| #> |71 |M **sep** 89 | 12| 0.6%| 1,727| 86.4%| #> |72 |M **sep** 93 | 12| 0.6%| 1,739| 87.0%| #> |73 |F **sep** 62 | 11| 0.6%| 1,750| 87.5%| #> |74 |F **sep** 64 | 11| 0.6%| 1,761| 88.1%| #> |75 |M **sep** 50 | 10| 0.5%| 1,771| 88.6%| #> |76 |F **sep** 43 | 9| 0.5%| 1,780| 89.0%| #> |77 |F **sep** 46 | 9| 0.5%| 1,789| 89.5%| #> |78 |F **sep** 51 | 9| 0.5%| 1,798| 89.9%| #> |79 |F **sep** 53 | 9| 0.5%| 1,807| 90.4%| #> |80 |F **sep** 55 | 9| 0.5%| 1,816| 90.8%| #> |81 |M **sep** 30 | 9| 0.5%| 1,825| 91.3%| #> |82 |F **sep** 44 | 8| 0.4%| 1,833| 91.7%| #> |83 |M **sep** 55 | 8| 0.4%| 1,841| 92.1%| #> |84 |F **sep** 41 | 7| 0.4%| 1,848| 92.4%| #> |85 |F **sep** 48 | 7| 0.4%| 1,855| 92.8%| #> |86 |F **sep** 52 | 7| 0.4%| 1,862| 93.1%| #> |87 |M **sep** 58 | 7| 0.4%| 1,869| 93.5%| #> |88 |M **sep** 60 | 7| 0.4%| 1,876| 93.8%| #> |89 |F **sep** 92 | 6| 0.3%| 1,882| 94.1%| #> |90 |M **sep** 43 | 6| 0.3%| 1,888| 94.4%| #> |91 |F **sep** 38 | 5| 0.3%| 1,893| 94.7%| #> |92 |F **sep** 42 | 5| 0.3%| 1,898| 94.9%| #> |93 |F **sep** 56 | 5| 0.3%| 1,903| 95.2%| #> |94 |M **sep** 44 | 5| 0.3%| 1,908| 95.4%| #> |95 |M **sep** 54 | 5| 0.3%| 1,913| 95.7%| #> |96 |F **sep** 20 | 4| 0.2%| 1,917| 95.9%| #> |97 |F **sep** 54 | 4| 0.2%| 1,921| 96.1%| #> |98 |F **sep** 61 | 4| 0.2%| 1,925| 96.3%| #> |99 |F **sep** 91 | 4| 0.2%| 1,929| 96.5%| #> |100 |F **sep** 93 | 4| 0.2%| 1,933| 96.7%| #> |101 |F **sep** 94 | 4| 0.2%| 1,937| 96.9%| #> |102 |M **sep** 46 | 4| 0.2%| 1,941| 97.1%| #> |103 |M **sep** 91 | 4| 0.2%| 1,945| 97.3%| #> |104 |F **sep** 39 | 3| 0.2%| 1,948| 97.4%| #> |105 |M **sep** 19 | 3| 0.2%| 1,951| 97.6%| #> |106 |M **sep** 33 | 3| 0.2%| 1,954| 97.7%| #> |107 |M **sep** 41 | 3| 0.2%| 1,957| 97.9%| #> |108 |M **sep** 48 | 3| 0.2%| 1,960| 98.0%| #> |109 |M **sep** 92 | 3| 0.2%| 1,963| 98.2%| #> |110 |F **sep** 31 | 2| 0.1%| 1,965| 98.3%| #> |111 |F **sep** 49 | 2| 0.1%| 1,967| 98.4%| #> |112 |M **sep** 18 | 2| 0.1%| 1,969| 98.5%| #> |113 |M **sep** 24 | 2| 0.1%| 1,971| 98.6%| #> |114 |M **sep** 29 | 2| 0.1%| 1,973| 98.7%| #> |115 |M **sep** 36 | 2| 0.1%| 1,975| 98.8%| #> |116 |M **sep** 37 | 2| 0.1%| 1,977| 98.9%| #> |117 |M **sep** 40 | 2| 0.1%| 1,979| 99.0%| #> |118 |M **sep** 42 | 2| 0.1%| 1,981| 99.1%| #> |119 |M **sep** 45 | 2| 0.1%| 1,983| 99.2%| #> |120 |M **sep** 49 | 2| 0.1%| 1,985| 99.3%| #> |121 |M **sep** 94 | 2| 0.1%| 1,987| 99.4%| #> |122 |M **sep** 97 | 2| 0.1%| 1,989| 99.5%| #> |123 |F **sep** 22 | 1| 0.1%| 1,990| 99.5%| #> |124 |F **sep** 29 | 1| 0.1%| 1,991| 99.6%| #> |125 |F **sep** 34 | 1| 0.1%| 1,992| 99.6%| #> |126 |F **sep** 35 | 1| 0.1%| 1,993| 99.7%| #> |127 |F **sep** 37 | 1| 0.1%| 1,994| 99.7%| #> |128 |F **sep** 40 | 1| 0.1%| 1,995| 99.8%| #> |129 |M **sep** 14 | 1| 0.1%| 1,996| 99.8%| #> |130 |M **sep** 32 | 1| 0.1%| 1,997| 99.9%| #> |131 |M **sep** 38 | 1| 0.1%| 1,998| 99.9%| #> |132 |M **sep** 39 | 1| 0.1%| 1,999| 100.0%| #> |133 |M **sep** 95 | 1| 0.1%| 2,000| 100.0%| #> #>#> #> #> **Frequency table of `hospital_id`** #> #> #> | |Item | Percent| #> |:--|:----|-------:| #> |1 |D | 38.1%| #> |2 |B | 33.2%| #> |3 |A | 16.1%| #> |4 |C | 12.7%| #> #>#> #> #> **Frequency table of `hospital_id`** #> #> #> | |Item | Percent| Cum. Percent| #> |:--|:----|-------:|------------:| #> |1 |D | 38.1%| 38.1%| #> |2 |B | 33.2%| 71.3%| #> |3 |A | 16.1%| 87.3%| #> |4 |C | 12.7%| 100.0%| #> #># check differences between frequency tables diff(freq(septic_patients$trim), freq(septic_patients$trsu))#> Differences between frequency tables #> #> |Item | Count #1| Count #2| Difference| Diff. percent| #> |:----|--------:|--------:|----------:|-------------:| #> |S | 918| 1392| +474| +51.6%| #> |R | 571| 361| -210| -36.8%| #> |I | 10| 6| -4| -40.0%|