NOTE: This page will be updated soon, as the pca() function is currently being developed.
=======NOTE: This page will be updated soon, as the pca() function is currently being developed.
>>>>>>> 8c9feea087f568fd4abbdb325140d1d628e6856fTransforming
<<<<<<< HEADFor PCA, we need to transform our AMR data first. This is what the
example_isolates data set in this package looks like:
For PCA, we need to transform our AMR data first. This is what the example_isolates data set in this package looks like:
library(AMR)
library(dplyr)
glimpse(example_isolates)
# Rows: 2,000
# Columns: 49
# $ date <date> 2002-01-02, 2002-01-03, 2002-01-07, 2002-01-07, 2002-…
# $ hospital_id <fct> D, D, B, B, B, B, D, D, B, B, D, D, D, D, D, B, B, B, …
# $ ward_icu <lgl> FALSE, FALSE, TRUE, TRUE, TRUE, TRUE, FALSE, FALSE, TR…
# $ ward_clinical <lgl> TRUE, TRUE, FALSE, FALSE, FALSE, FALSE, TRUE, TRUE, FA…
# $ ward_outpatient <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE…
# $ age <dbl> 65, 65, 45, 45, 45, 45, 78, 78, 45, 79, 67, 67, 71, 71…
# $ gender <chr> "F", "F", "F", "F", "F", "F", "M", "M", "F", "F", "M",…
# $ patient_id <chr> "A77334", "A77334", "067927", "067927", "067927", "067…
# $ mo <mo> "B_ESCHR_COLI", "B_ESCHR_COLI", "B_STPHY_EPDR", "B_STPH…
# $ PEN <rsi> R, R, R, R, R, R, R, R, R, R, R, R, R, R, R, R, R, R, …
# $ OXA <rsi> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
# $ FLC <rsi> NA, NA, R, R, R, R, S, S, R, S, S, S, NA, NA, NA, NA, …
# $ AMX <rsi> NA, NA, NA, NA, NA, NA, R, R, NA, NA, NA, NA, NA, NA, …
# $ AMC <rsi> I, I, NA, NA, NA, NA, S, S, NA, NA, S, S, I, I, R, I, …
# $ AMP <rsi> NA, NA, NA, NA, NA, NA, R, R, NA, NA, NA, NA, NA, NA, …
# $ TZP <rsi> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
# $ CZO <rsi> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
# $ FEP <rsi> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
# $ CXM <rsi> I, I, R, R, R, R, S, S, R, S, S, S, S, S, NA, S, S, R,…
# $ FOX <rsi> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
# $ CTX <rsi> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, S, S, …
# $ CAZ <rsi> NA, NA, R, R, R, R, R, R, R, R, R, R, NA, NA, NA, S, S…
# $ CRO <rsi> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, S, S, …
# $ GEN <rsi> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
# $ TOB <rsi> NA, NA, NA, NA, NA, NA, S, S, NA, NA, NA, NA, S, S, NA…
# $ AMK <rsi> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
# $ KAN <rsi> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
# $ TMP <rsi> R, R, S, S, R, R, R, R, S, S, NA, NA, S, S, S, S, S, R…
# $ SXT <rsi> R, R, S, S, NA, NA, NA, NA, S, S, NA, NA, S, S, S, S, …
# $ NIT <rsi> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
# $ FOS <rsi> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
# $ LNZ <rsi> R, R, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, R, R, R,…
# $ CIP <rsi> NA, NA, NA, NA, NA, NA, NA, NA, S, S, NA, NA, NA, NA, …
# $ MFX <rsi> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
# $ VAN <rsi> R, R, S, S, S, S, S, S, S, S, NA, NA, R, R, R, R, R, S…
# $ TEC <rsi> R, R, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, R, R, R,…
# $ TCY <rsi> R, R, S, S, S, S, S, S, S, I, S, S, NA, NA, I, R, R, S…
# $ TGC <rsi> NA, NA, S, S, S, S, S, S, S, NA, S, S, NA, NA, NA, R, …
# $ DOX <rsi> NA, NA, S, S, S, S, S, S, S, NA, S, S, NA, NA, NA, R, …
# $ ERY <rsi> R, R, R, R, R, R, S, S, R, S, S, S, R, R, R, R, R, R, …
# $ CLI <rsi> R, R, NA, NA, NA, R, NA, NA, NA, NA, NA, NA, R, R, R, …
# $ AZM <rsi> R, R, R, R, R, R, S, S, R, S, S, S, R, R, R, R, R, R, …
# $ IPM <rsi> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, S, S, …
# $ MEM <rsi> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
# $ MTR <rsi> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
# $ CHL <rsi> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
# $ COL <rsi> NA, NA, R, R, R, R, R, R, R, R, R, R, NA, NA, NA, R, R…
# $ MUP <rsi> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
# $ RIF <rsi> R, R, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, R, R, R,…Now to transform this to a data set with only resistance percentages per taxonomic order and genus:
resistance_data <- example_isolates %>%
group_by(order = mo_order(mo), # group on anything, like order
genus = mo_genus(mo)) %>% # and genus as we do here
summarise_if(is.rsi, resistance) %>% # then get resistance of all drugs
select(order, genus, AMC, CXM, CTX,
CAZ, GEN, TOB, TMP, SXT) # and select only relevant columns
head(resistance_data)
# # A tibble: 6 × 10
# # Groups: order [5]
# order genus AMC CXM CTX CAZ GEN TOB TMP SXT
# <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
# 1 (unknown order) (unknown ge… NA NA NA NA NA NA NA NA
# 2 Actinomycetales Schaalia NA NA NA NA NA NA NA NA
# 3 Bacteroidales Bacteroides NA NA NA NA NA NA NA NA
# 4 Campylobacterales Campylobact… NA NA NA NA NA NA NA NA
# 5 Caryophanales Gemella NA NA NA NA NA NA NA NA
# 6 Caryophanales Listeria NA NA NA NA NA NA NA NAPerform principal component analysis
<<<<<<< HEADThe new pca() function will automatically filter on rows
that contain numeric values in all selected variables, so we now only
need to do:
The new pca() function will automatically filter on rows that contain numeric values in all selected variables, so we now only need to do:
pca_result <- pca(resistance_data)
# ℹ Columns selected for PCA: "AMC", "CAZ", "CTX", "CXM", "GEN", "SXT", "TMP"
# and "TOB". Total observations available: 7.The result can be reviewed with the good old summary()
function:
summary(pca_result)
# Groups (n=4, named as 'order'):
# [1] "Caryophanales" "Enterobacterales" "Lactobacillales" "Pseudomonadales"
# Importance of components:
# PC1 PC2 PC3 PC4 PC5 PC6 PC7
# Standard deviation 2.1539 1.6807 0.6138 0.33879 0.20808 0.03140 5.121e-17
# Proportion of Variance 0.5799 0.3531 0.0471 0.01435 0.00541 0.00012 0.000e+00
# Cumulative Proportion 0.5799 0.9330 0.9801 0.99446 0.99988 1.00000 1.000e+00# Groups (n=4, named as 'order'):
# [1] "Caryophanales" "Enterobacterales" "Lactobacillales" "Pseudomonadales"Good news. The first two components explain a total of 93.3% of the
variance (see the PC1 and PC2 values of the Proportion of
Variance. We can create a so-called biplot with the base R
biplot() function, to see which antimicrobial resistance
per drug explain the difference per microorganism.
Plotting the results
biplot(pca_result)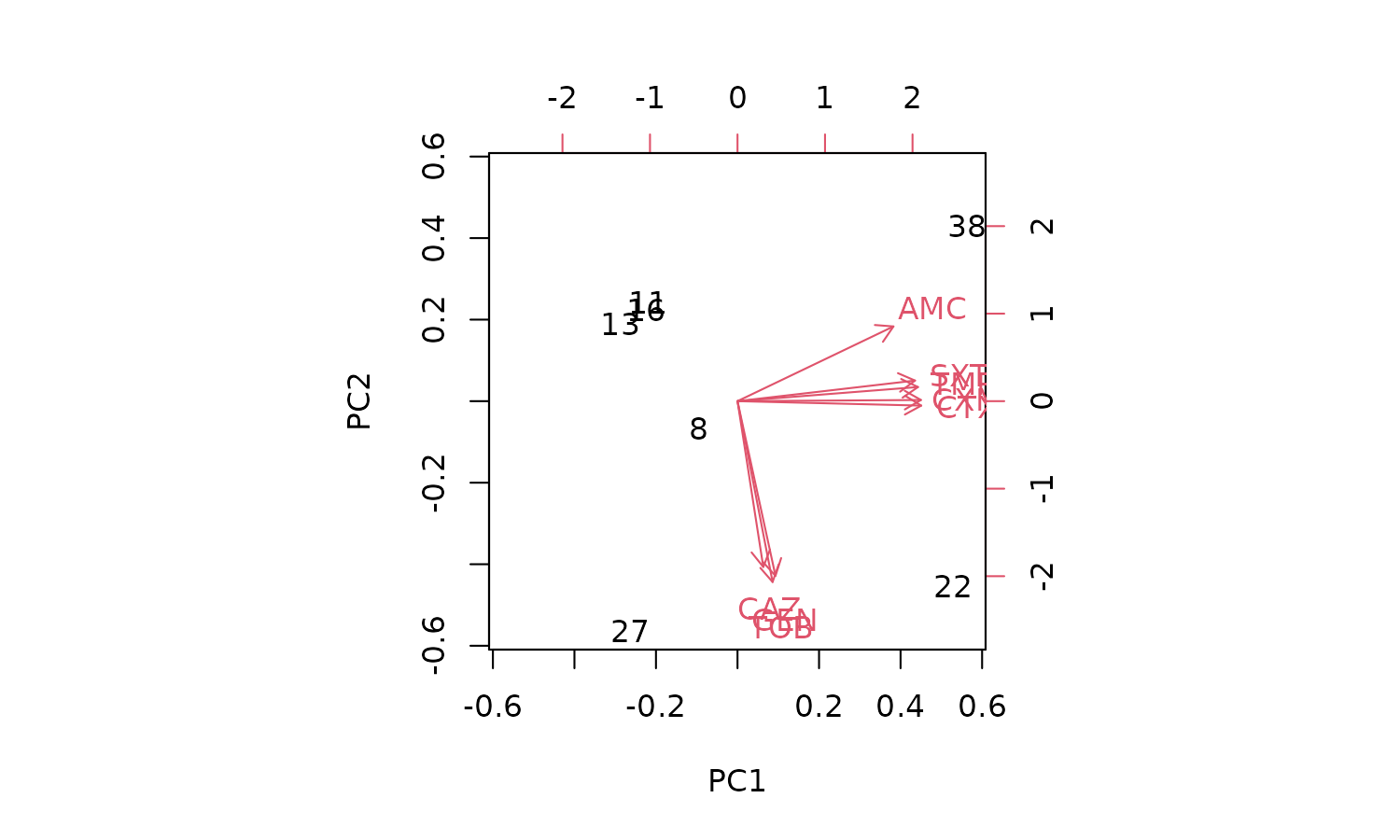
But we can’t see the explanation of the points. Perhaps this works
better with our new ggplot_pca() function, that
automatically adds the right labels and even groups:
ggplot_pca(pca_result)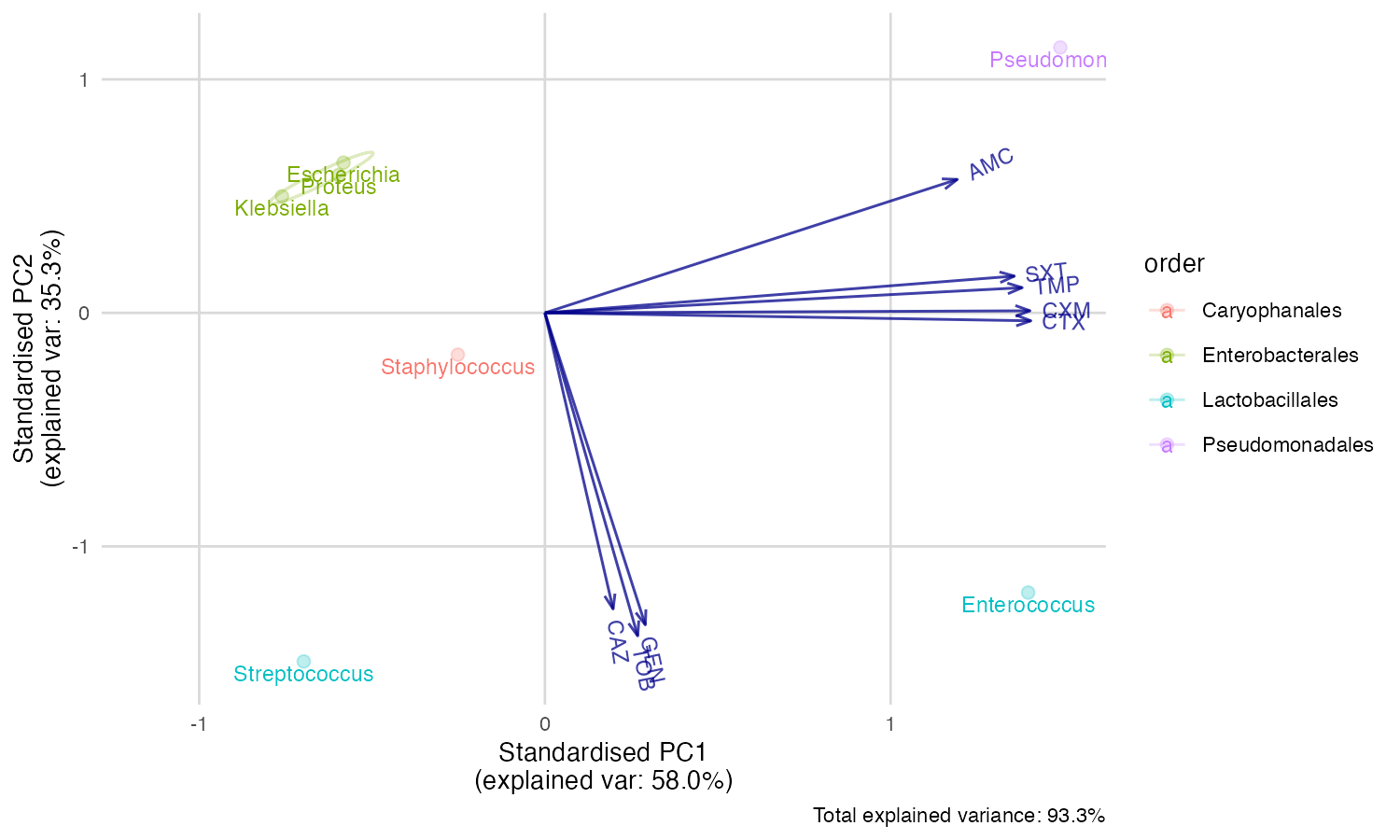
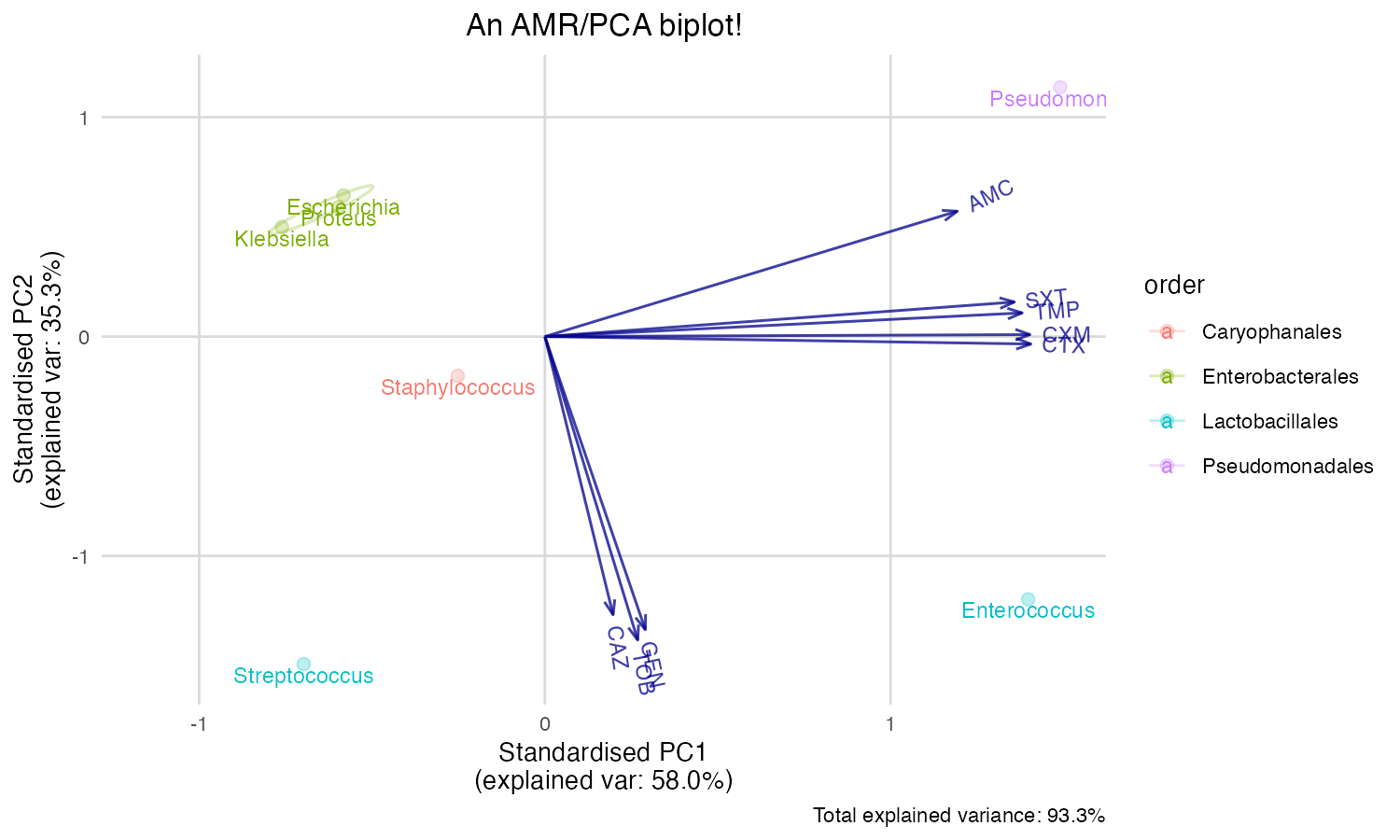
You can also print an ellipse per group, and edit the appearance:
ggplot_pca(pca_result, ellipse = TRUE) +
ggplot2::labs(title = "An AMR/PCA biplot!")