Performs a principal component analysis (PCA) based on a data set with automatic determination for afterwards plotting the groups and labels, and automatic filtering on only suitable (i.e. non-empty and numeric) variables.
Arguments
- x
a data.frame containing numeric columns
- ...
columns of
xto be selected for PCA, can be unquoted since it supports quasiquotation.- retx
a logical value indicating whether the rotated variables should be returned.
- center
a logical value indicating whether the variables should be shifted to be zero centered. Alternately, a vector of length equal the number of columns of
xcan be supplied. The value is passed toscale.- scale.
a logical value indicating whether the variables should be scaled to have unit variance before the analysis takes place. The default is
FALSEfor consistency with S, but in general scaling is advisable. Alternatively, a vector of length equal the number of columns ofxcan be supplied. The value is passed toscale.- tol
a value indicating the magnitude below which components should be omitted. (Components are omitted if their standard deviations are less than or equal to
toltimes the standard deviation of the first component.) With the default null setting, no components are omitted (unlessrank.is specified less thanmin(dim(x)).). Other settings for tol could betol = 0ortol = sqrt(.Machine$double.eps), which would omit essentially constant components.- rank.
optionally, a number specifying the maximal rank, i.e., maximal number of principal components to be used. Can be set as alternative or in addition to
tol, useful notably when the desired rank is considerably smaller than the dimensions of the matrix.
Value
An object of classes pca and prcomp
Details
The pca() function takes a data.frame as input and performs the actual PCA with the R function prcomp().
The result of the pca() function is a prcomp object, with an additional attribute non_numeric_cols which is a vector with the column names of all columns that do not contain numeric values. These are probably the groups and labels, and will be used by ggplot_pca().
Examples
# `example_isolates` is a data set available in the AMR package.
# See ?example_isolates.
# \donttest{
if (require("dplyr")) {
# calculate the resistance per group first
resistance_data <- example_isolates %>%
group_by(
order = mo_order(mo), # group on anything, like order
genus = mo_genus(mo)
) %>% # and genus as we do here;
filter(n() >= 30) %>% # filter on only 30 results per group
summarise_if(is.rsi, resistance) # then get resistance of all drugs
# now conduct PCA for certain antimicrobial agents
pca_result <- resistance_data %>%
pca(AMC, CXM, CTX, CAZ, GEN, TOB, TMP, SXT)
pca_result
summary(pca_result)
# old base R plotting method:
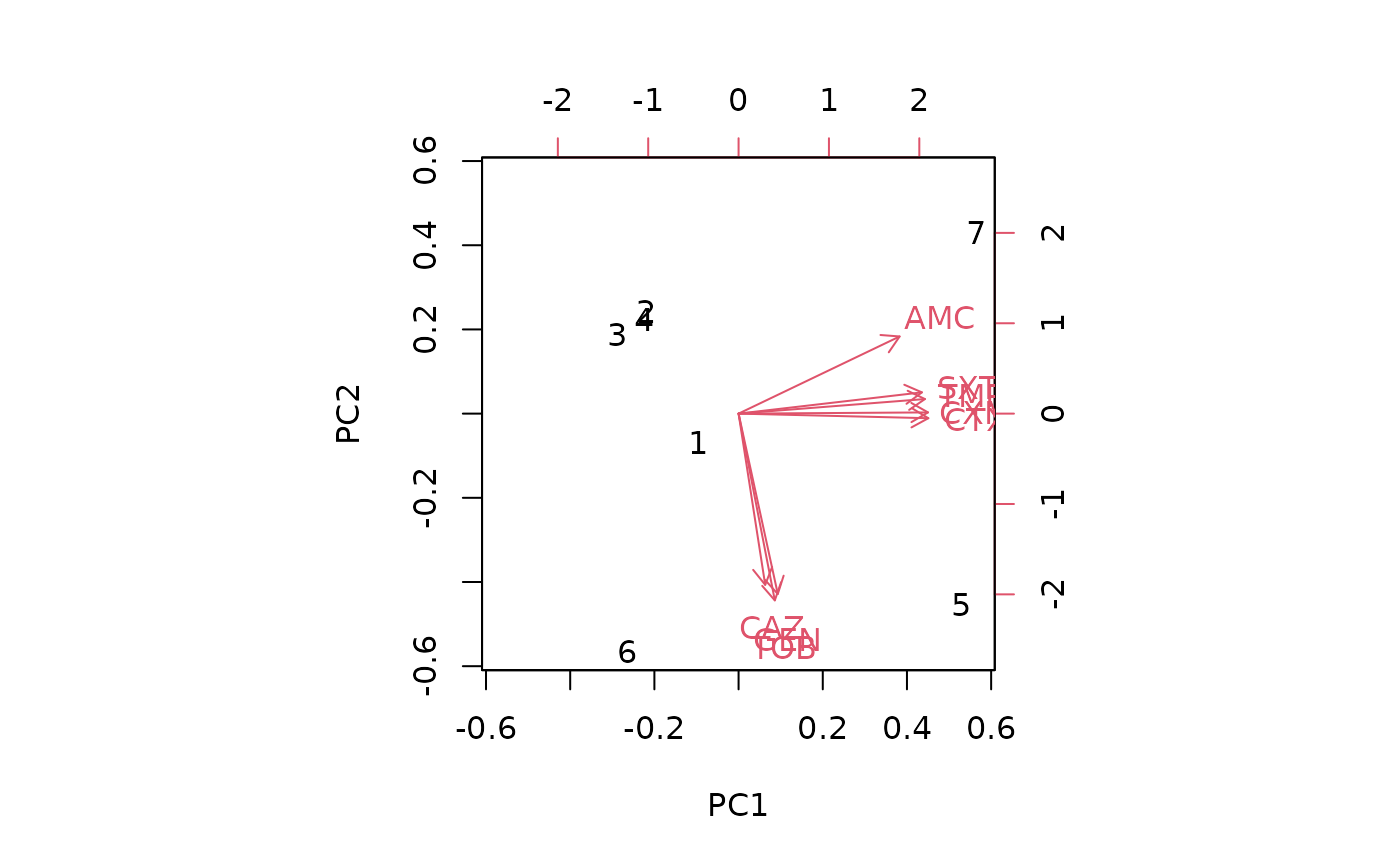
biplot(pca_result)
# new ggplot2 plotting method using this package:
if (require("ggplot2")) {
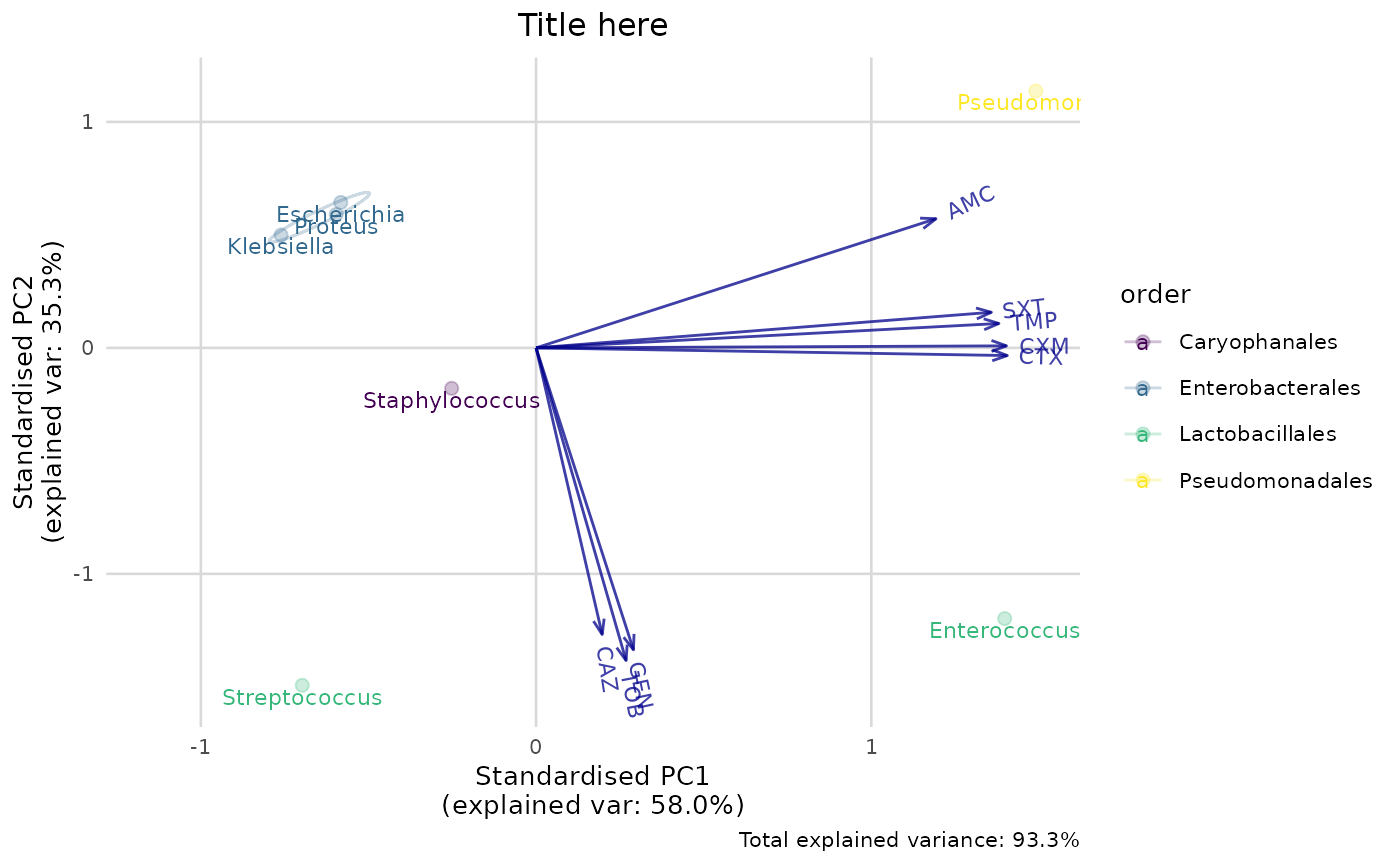
ggplot_pca(pca_result)
ggplot_pca(pca_result) +
scale_colour_viridis_d() +
labs(title = "Title here")
}
}
#> Warning: Introducing NA: only 14 results available for PEN in group: order =
#> "Lactobacillales", genus = "Enterococcus" (`minimum` = 30).
#> Warning: Introducing NA: no results available for OXA in group: order =
#> "Enterobacterales", genus = "Escherichia" (`minimum` = 30).
#> Warning: Introducing NA: no results available for OXA in group: order =
#> "Enterobacterales", genus = "Klebsiella" (`minimum` = 30).
#> Warning: Introducing NA: no results available for OXA in group: order =
#> "Enterobacterales", genus = "Proteus" (`minimum` = 30).
#> Warning: Introducing NA: only 13 results available for OXA in group: order =
#> "Lactobacillales", genus = "Enterococcus" (`minimum` = 30).
#> Warning: Introducing NA: only 15 results available for OXA in group: order =
#> "Lactobacillales", genus = "Streptococcus" (`minimum` = 30).
#> Warning: Introducing NA: no results available for OXA in group: order =
#> "Pseudomonadales", genus = "Pseudomonas" (`minimum` = 30).
#> Warning: Introducing NA: no results available for FLC in group: order =
#> "Enterobacterales", genus = "Escherichia" (`minimum` = 30).
#> Warning: Introducing NA: no results available for FLC in group: order =
#> "Enterobacterales", genus = "Klebsiella" (`minimum` = 30).
#> Warning: Introducing NA: no results available for FLC in group: order =
#> "Enterobacterales", genus = "Proteus" (`minimum` = 30).
#> Warning: Introducing NA: only 13 results available for FLC in group: order =
#> "Lactobacillales", genus = "Enterococcus" (`minimum` = 30).
#> Warning: Introducing NA: no results available for FLC in group: order =
#> "Pseudomonadales", genus = "Pseudomonas" (`minimum` = 30).
#> Warning: Introducing NA: only 26 results available for AMX in group: order =
#> "Enterobacterales", genus = "Proteus" (`minimum` = 30).
#> Warning: Introducing NA: only 26 results available for AMP in group: order =
#> "Enterobacterales", genus = "Proteus" (`minimum` = 30).
#> Warning: Introducing NA: only 27 results available for TZP in group: order =
#> "Pseudomonadales", genus = "Pseudomonas" (`minimum` = 30).
#> Warning: Introducing NA: only 12 results available for CZO in group: order =
#> "Enterobacterales", genus = "Klebsiella" (`minimum` = 30).
#> Warning: Introducing NA: only 5 results available for CZO in group: order =
#> "Enterobacterales", genus = "Proteus" (`minimum` = 30).
#> Warning: Introducing NA: only 27 results available for FEP in group: order =
#> "Enterobacterales", genus = "Proteus" (`minimum` = 30).
#> Warning: Introducing NA: only 23 results available for FEP in group: order =
#> "Pseudomonadales", genus = "Pseudomonas" (`minimum` = 30).
#> Warning: Introducing NA: only 29 results available for FOX in group: order =
#> "Enterobacterales", genus = "Proteus" (`minimum` = 30).
#> Warning: Introducing NA: only 26 results available for AMK in group: order =
#> "Enterobacterales", genus = "Klebsiella" (`minimum` = 30).
#> Warning: Introducing NA: only 6 results available for AMK in group: order =
#> "Enterobacterales", genus = "Proteus" (`minimum` = 30).
#> Warning: Introducing NA: only 17 results available for AMK in group: order =
#> "Pseudomonadales", genus = "Pseudomonas" (`minimum` = 30).
#> Warning: Introducing NA: no results available for KAN in group: order =
#> "Enterobacterales", genus = "Escherichia" (`minimum` = 30).
#> Warning: Introducing NA: no results available for KAN in group: order =
#> "Enterobacterales", genus = "Klebsiella" (`minimum` = 30).
#> Warning: Introducing NA: no results available for KAN in group: order =
#> "Enterobacterales", genus = "Proteus" (`minimum` = 30).
#> Warning: Introducing NA: only 6 results available for NIT in group: order =
#> "Lactobacillales", genus = "Streptococcus" (`minimum` = 30).
#> Warning: Introducing NA: only 17 results available for NIT in group: order =
#> "Pseudomonadales", genus = "Pseudomonas" (`minimum` = 30).
#> Warning: Introducing NA: only 8 results available for FOS in group: order =
#> "Enterobacterales", genus = "Klebsiella" (`minimum` = 30).
#> Warning: Introducing NA: only 2 results available for FOS in group: order =
#> "Enterobacterales", genus = "Proteus" (`minimum` = 30).
#> Warning: Introducing NA: no results available for FOS in group: order =
#> "Lactobacillales", genus = "Enterococcus" (`minimum` = 30).
#> Warning: Introducing NA: no results available for FOS in group: order =
#> "Lactobacillales", genus = "Streptococcus" (`minimum` = 30).
#> Warning: Introducing NA: no results available for FOS in group: order =
#> "Pseudomonadales", genus = "Pseudomonas" (`minimum` = 30).
#> Warning: Introducing NA: only 7 results available for LNZ in group: order =
#> "Lactobacillales", genus = "Streptococcus" (`minimum` = 30).
#> Warning: Introducing NA: only 5 results available for CIP in group: order =
#> "Lactobacillales", genus = "Enterococcus" (`minimum` = 30).
#> Warning: Introducing NA: only 23 results available for CIP in group: order =
#> "Lactobacillales", genus = "Streptococcus" (`minimum` = 30).
#> Warning: Introducing NA: only 2 results available for MFX in group: order =
#> "Enterobacterales", genus = "Klebsiella" (`minimum` = 30).
#> Warning: Introducing NA: only 2 results available for MFX in group: order =
#> "Enterobacterales", genus = "Proteus" (`minimum` = 30).
#> Warning: Introducing NA: no results available for MFX in group: order =
#> "Lactobacillales", genus = "Enterococcus" (`minimum` = 30).
#> Warning: Introducing NA: only 7 results available for MFX in group: order =
#> "Lactobacillales", genus = "Streptococcus" (`minimum` = 30).
#> Warning: Introducing NA: no results available for MFX in group: order =
#> "Pseudomonadales", genus = "Pseudomonas" (`minimum` = 30).
#> Warning: Introducing NA: only 6 results available for TEC in group: order =
#> "Lactobacillales", genus = "Streptococcus" (`minimum` = 30).
#> Warning: Introducing NA: only 3 results available for TCY in group: order =
#> "Enterobacterales", genus = "Escherichia" (`minimum` = 30).
#> Warning: Introducing NA: only 2 results available for TCY in group: order =
#> "Enterobacterales", genus = "Klebsiella" (`minimum` = 30).
#> Warning: Introducing NA: only 18 results available for TGC in group: order =
#> "Enterobacterales", genus = "Klebsiella" (`minimum` = 30).
#> Warning: Introducing NA: only 7 results available for TGC in group: order =
#> "Lactobacillales", genus = "Streptococcus" (`minimum` = 30).
#> Warning: Introducing NA: no results available for DOX in group: order =
#> "Enterobacterales", genus = "Escherichia" (`minimum` = 30).
#> Warning: Introducing NA: no results available for DOX in group: order =
#> "Enterobacterales", genus = "Klebsiella" (`minimum` = 30).
#> Warning: Introducing NA: only 2 results available for DOX in group: order =
#> "Lactobacillales", genus = "Enterococcus" (`minimum` = 30).
#> Warning: Introducing NA: only 27 results available for IPM in group: order =
#> "Pseudomonadales", genus = "Pseudomonas" (`minimum` = 30).
#> Warning: Introducing NA: only 25 results available for MEM in group: order =
#> "Lactobacillales", genus = "Enterococcus" (`minimum` = 30).
#> Warning: Introducing NA: only 26 results available for MEM in group: order =
#> "Pseudomonadales", genus = "Pseudomonas" (`minimum` = 30).
#> Warning: Introducing NA: no results available for MTR in group: order =
#> "Caryophanales", genus = "Staphylococcus" (`minimum` = 30).
#> Warning: Introducing NA: only 2 results available for MTR in group: order =
#> "Enterobacterales", genus = "Escherichia" (`minimum` = 30).
#> Warning: Introducing NA: no results available for MTR in group: order =
#> "Enterobacterales", genus = "Klebsiella" (`minimum` = 30).
#> Warning: Introducing NA: no results available for MTR in group: order =
#> "Enterobacterales", genus = "Proteus" (`minimum` = 30).
#> Warning: Introducing NA: no results available for MTR in group: order =
#> "Lactobacillales", genus = "Enterococcus" (`minimum` = 30).
#> Warning: Introducing NA: no results available for MTR in group: order =
#> "Lactobacillales", genus = "Streptococcus" (`minimum` = 30).
#> Warning: Introducing NA: no results available for MTR in group: order =
#> "Pseudomonadales", genus = "Pseudomonas" (`minimum` = 30).
#> Warning: Introducing NA: no results available for CHL in group: order =
#> "Enterobacterales", genus = "Escherichia" (`minimum` = 30).
#> Warning: Introducing NA: no results available for CHL in group: order =
#> "Enterobacterales", genus = "Klebsiella" (`minimum` = 30).
#> Warning: Introducing NA: no results available for CHL in group: order =
#> "Enterobacterales", genus = "Proteus" (`minimum` = 30).
#> Warning: Introducing NA: no results available for CHL in group: order =
#> "Lactobacillales", genus = "Enterococcus" (`minimum` = 30).
#> Warning: Introducing NA: no results available for CHL in group: order =
#> "Lactobacillales", genus = "Streptococcus" (`minimum` = 30).
#> Warning: Introducing NA: only 9 results available for COL in group: order =
#> "Pseudomonadales", genus = "Pseudomonas" (`minimum` = 30).
#> Warning: Introducing NA: no results available for MUP in group: order =
#> "Enterobacterales", genus = "Escherichia" (`minimum` = 30).
#> Warning: Introducing NA: no results available for MUP in group: order =
#> "Enterobacterales", genus = "Klebsiella" (`minimum` = 30).
#> Warning: Introducing NA: no results available for MUP in group: order =
#> "Enterobacterales", genus = "Proteus" (`minimum` = 30).
#> Warning: Introducing NA: no results available for MUP in group: order =
#> "Lactobacillales", genus = "Enterococcus" (`minimum` = 30).
#> Warning: Introducing NA: no results available for MUP in group: order =
#> "Lactobacillales", genus = "Streptococcus" (`minimum` = 30).
#> Warning: Introducing NA: no results available for MUP in group: order =
#> "Pseudomonadales", genus = "Pseudomonas" (`minimum` = 30).
#> Warning: Introducing NA: no results available for RIF in group: order =
#> "Lactobacillales", genus = "Enterococcus" (`minimum` = 30).
#> Warning: Introducing NA: no results available for RIF in group: order =
#> "Lactobacillales", genus = "Streptococcus" (`minimum` = 30).
#> ℹ Columns selected for PCA: "AMC", "CAZ", "CTX", "CXM", "GEN", "SXT", "TMP"
#> and "TOB". Total observations available: 7.
#> Groups (n=4, named as 'order'):
#> [1] "Caryophanales" "Enterobacterales" "Lactobacillales" "Pseudomonadales"
#>
 # }
# }