Calculate resistance of isolates
portion.RdThese functions can be used to calculate the (co-)resistance of microbial isolates (i.e. percentage of S, SI, I, IR or R). All functions support quasiquotation with pipes, can be used in dplyrs summarise and support grouped variables, see Examples.
portion_R and portion_IR can be used to calculate resistance, portion_S and portion_SI can be used to calculate susceptibility.
portion_R(..., minimum = 30, as_percent = FALSE, also_single_tested = FALSE) portion_IR(..., minimum = 30, as_percent = FALSE, also_single_tested = FALSE) portion_I(..., minimum = 30, as_percent = FALSE, also_single_tested = FALSE) portion_SI(..., minimum = 30, as_percent = FALSE, also_single_tested = FALSE) portion_S(..., minimum = 30, as_percent = FALSE, also_single_tested = FALSE) portion_df(data, translate_ab = "name", language = get_locale(), minimum = 30, as_percent = FALSE, combine_SI = TRUE, combine_IR = FALSE)
Arguments
| ... | one or more vectors (or columns) with antibiotic interpretations. They will be transformed internally with |
|---|---|
| minimum | the minimum allowed number of available (tested) isolates. Any isolate count lower than |
| as_percent | a logical to indicate whether the output must be returned as a hundred fold with % sign (a character). A value of |
| also_single_tested | a logical to indicate whether (in combination therapies) also observations should be included where not all antibiotics were tested, but at least one of the tested antibiotics contains a target interpretation (e.g. S in case of |
| data | a |
| translate_ab | a column name of the |
| language | language of the returned text, defaults to system language (see |
| combine_SI | a logical to indicate whether all values of S and I must be merged into one, so the output only consists of S+I vs. R (susceptible vs. resistant). This used to be the parameter |
| combine_IR | a logical to indicate whether all values of I and R must be merged into one, so the output only consists of S vs. I+R (susceptible vs. non-susceptible). This is outdated, see parameter |
Source
M39 Analysis and Presentation of Cumulative Antimicrobial Susceptibility Test Data, 4th Edition, 2014, Clinical and Laboratory Standards Institute (CLSI). https://clsi.org/standards/products/microbiology/documents/m39/.
Wickham H. Tidy Data. The Journal of Statistical Software, vol. 59, 2014. http://vita.had.co.nz/papers/tidy-data.html
Value
Double or, when as_percent = TRUE, a character.
Details
Remember that you should filter your table to let it contain only first isolates! Use first_isolate to determine them in your data set.
These functions are not meant to count isolates, but to calculate the portion of resistance/susceptibility. Use the count functions to count isolates. Low counts can infuence the outcome - these portion functions may camouflage this, since they only return the portion albeit being dependent on the minimum parameter.
portion_df takes any variable from data that has an "rsi" class (created with as.rsi) and calculates the portions R, I and S. The resulting tidy data (see Source) data.frame will have three rows (S/I/R) and a column for each variable with class "rsi".
To calculate the probability (p) of susceptibility of one antibiotic, we use this formula:
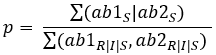
For two antibiotics:
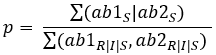
For three antibiotics:
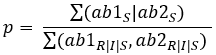
And so on.
Interpretation of S, I and R
In 2019, EUCAST has decided to change the definitions of susceptibility testing categories S, I and R as shown below. Results of several consultations on the new definitions are available on the EUCAST website under "Consultations".
S - Susceptible, standard dosing regimen: A microorganism is categorised as "Susceptible, standard dosing regimen", when there is a high likelihood of therapeutic success using a standard dosing regimen of the agent.
I - Susceptible, increased exposure: A microorganism is categorised as "Susceptible, Increased exposure" when there is a high likelihood of therapeutic success because exposure to the agent is increased by adjusting the dosing regimen or by its concentration at the site of infection.
R - Resistant: A microorganism is categorised as "Resistant" when there is a high likelihood of therapeutic failure even when there is increased exposure.
Exposure is a function of how the mode of administration, dose, dosing interval, infusion time, as well as distribution and excretion of the antimicrobial agent will influence the infecting organism at the site of infection.
Source: http://www.eucast.org/newsiandr/.
This AMR package honours this new insight.
Read more on our website!
On our website https://msberends.gitlab.io/AMR you can find a tutorial about how to conduct AMR analysis, the complete documentation of all functions (which reads a lot easier than here in R) and an example analysis using WHONET data.
See also
count_* to count resistant and susceptible isolates.
Examples
# NOT RUN { # septic_patients is a data set available in the AMR package. It is true, genuine data. ?septic_patients # Calculate resistance portion_R(septic_patients$AMX) portion_IR(septic_patients$AMX) # Or susceptibility portion_S(septic_patients$AMX) portion_SI(septic_patients$AMX) # Do the above with pipes: library(dplyr) septic_patients %>% portion_R(AMX) septic_patients %>% portion_IR(AMX) septic_patients %>% portion_S(AMX) septic_patients %>% portion_SI(AMX) septic_patients %>% group_by(hospital_id) %>% summarise(p = portion_S(CIP), n = n_rsi(CIP)) # n_rsi works like n_distinct in dplyr septic_patients %>% group_by(hospital_id) %>% summarise(R = portion_R(CIP, as_percent = TRUE), I = portion_I(CIP, as_percent = TRUE), S = portion_S(CIP, as_percent = TRUE), n1 = count_all(CIP), # the actual total; sum of all three n2 = n_rsi(CIP), # same - analogous to n_distinct total = n()) # NOT the number of tested isolates! # Calculate co-resistance between amoxicillin/clav acid and gentamicin, # so we can see that combination therapy does a lot more than mono therapy: septic_patients %>% portion_S(AMC) # S = 71.4% septic_patients %>% count_all(AMC) # n = 1879 septic_patients %>% portion_S(GEN) # S = 74.0% septic_patients %>% count_all(GEN) # n = 1855 septic_patients %>% portion_S(AMC, GEN) # S = 92.3% septic_patients %>% count_all(AMC, GEN) # n = 1798 septic_patients %>% group_by(hospital_id) %>% summarise(cipro_p = portion_S(CIP, as_percent = TRUE), cipro_n = count_all(CIP), genta_p = portion_S(GEN, as_percent = TRUE), genta_n = count_all(GEN), combination_p = portion_S(CIP, GEN, as_percent = TRUE), combination_n = count_all(CIP, GEN)) # Get portions S/I/R immediately of all rsi columns septic_patients %>% select(AMX, CIP) %>% portion_df(translate = FALSE) # It also supports grouping variables septic_patients %>% select(hospital_id, AMX, CIP) %>% group_by(hospital_id) %>% portion_df(translate = FALSE) # }# NOT RUN { # calculate current empiric combination therapy of Helicobacter gastritis: my_table %>% filter(first_isolate == TRUE, genus == "Helicobacter") %>% summarise(p = portion_S(AMX, MTR), # amoxicillin with metronidazole n = count_all(AMX, MTR)) # }