Use these functions to return a specific property of a microorganism based on the latest accepted taxonomy. All input values will be evaluated internally with as.mo(), which makes it possible to use microbial abbreviations, codes and names as input. See Examples.
Usage
mo_name(x, language = get_AMR_locale(), ...)
mo_fullname(x, language = get_AMR_locale(), ...)
mo_shortname(x, language = get_AMR_locale(), ...)
mo_subspecies(x, language = get_AMR_locale(), ...)
mo_species(x, language = get_AMR_locale(), ...)
mo_genus(x, language = get_AMR_locale(), ...)
mo_family(x, language = get_AMR_locale(), ...)
mo_order(x, language = get_AMR_locale(), ...)
mo_class(x, language = get_AMR_locale(), ...)
mo_phylum(x, language = get_AMR_locale(), ...)
mo_kingdom(x, language = get_AMR_locale(), ...)
mo_domain(x, language = get_AMR_locale(), ...)
mo_type(x, language = get_AMR_locale(), ...)
mo_gramstain(x, language = get_AMR_locale(), ...)
mo_is_gram_negative(x, language = get_AMR_locale(), ...)
mo_is_gram_positive(x, language = get_AMR_locale(), ...)
mo_is_yeast(x, language = get_AMR_locale(), ...)
mo_is_intrinsic_resistant(x, ab, language = get_AMR_locale(), ...)
mo_snomed(x, language = get_AMR_locale(), ...)
mo_ref(x, language = get_AMR_locale(), ...)
mo_authors(x, language = get_AMR_locale(), ...)
mo_year(x, language = get_AMR_locale(), ...)
mo_lpsn(x, language = get_AMR_locale(), ...)
mo_rank(x, language = get_AMR_locale(), ...)
mo_taxonomy(x, language = get_AMR_locale(), ...)
mo_synonyms(x, language = get_AMR_locale(), ...)
mo_info(x, language = get_AMR_locale(), ...)
mo_url(x, open = FALSE, language = get_AMR_locale(), ...)
mo_property(x, property = "fullname", language = get_AMR_locale(), ...)Arguments
- x
any character (vector) that can be coerced to a valid microorganism code with
as.mo(). Can be left blank for auto-guessing the column containing microorganism codes if used in a data set, see Examples.- language
language of the returned text, defaults to system language (see
get_AMR_locale()) and can be overwritten by setting the optionAMR_locale, e.g.options(AMR_locale = "de"), see translate. Also used to translate text like "no growth". Uselanguage = NULLorlanguage = ""to prevent translation.- ...
other arguments passed on to
as.mo(), such as 'allow_uncertain' and 'ignore_pattern'- ab
any (vector of) text that can be coerced to a valid antibiotic code with
as.ab()- open
browse the URL using
browseURL()- property
one of the column names of the microorganisms data set: "mo", "fullname", "kingdom", "phylum", "class", "order", "family", "genus", "species", "subspecies", "rank", "ref", "species_id", "source", "prevalence" or "snomed", or must be
"shortname"
Details
All functions will return the most recently known taxonomic property according to the Catalogue of Life, except for mo_ref(), mo_authors() and mo_year(). Please refer to this example, knowing that Escherichia blattae was renamed to Shimwellia blattae in 2010:
mo_name("Escherichia blattae")will return"Shimwellia blattae"(with a message about the renaming)mo_ref("Escherichia blattae")will return"Burgess et al., 1973"(with a message about the renaming)mo_ref("Shimwellia blattae")will return"Priest et al., 2010"(without a message)
The short name - mo_shortname() - almost always returns the first character of the genus and the full species, like "E. coli". Exceptions are abbreviations of staphylococci (such as "CoNS", Coagulase-Negative Staphylococci) and beta-haemolytic streptococci (such as "GBS", Group B Streptococci). Please bear in mind that e.g. E. coli could mean Escherichia coli (kingdom of Bacteria) as well as Entamoeba coli (kingdom of Protozoa). Returning to the full name will be done using as.mo() internally, giving priority to bacteria and human pathogens, i.e. "E. coli" will be considered Escherichia coli. In other words, mo_fullname(mo_shortname("Entamoeba coli")) returns "Escherichia coli".
Since the top-level of the taxonomy is sometimes referred to as 'kingdom' and sometimes as 'domain', the functions mo_kingdom() and mo_domain() return the exact same results.
The Gram stain - mo_gramstain() - will be determined based on the taxonomic kingdom and phylum. According to Cavalier-Smith (2002, PMID 11837318), who defined subkingdoms Negibacteria and Posibacteria, only these phyla are Posibacteria: Actinobacteria, Chloroflexi, Firmicutes and Tenericutes. These bacteria are considered Gram-positive, except for members of the class Negativicutes which are Gram-negative. Members of other bacterial phyla are all considered Gram-negative. Species outside the kingdom of Bacteria will return a value NA. Functions mo_is_gram_negative() and mo_is_gram_positive() always return TRUE or FALSE (except when the input is NA or the MO code is UNKNOWN), thus always return FALSE for species outside the taxonomic kingdom of Bacteria.
Determination of yeasts - mo_is_yeast() - will be based on the taxonomic kingdom and class. Budding yeasts are fungi of the phylum Ascomycetes, class Saccharomycetes (also called Hemiascomycetes). True yeasts are aggregated into the underlying order Saccharomycetales. Thus, for all microorganisms that are fungi and member of the taxonomic class Saccharomycetes, the function will return TRUE. It returns FALSE otherwise (except when the input is NA or the MO code is UNKNOWN).
Intrinsic resistance - mo_is_intrinsic_resistant() - will be determined based on the intrinsic_resistant data set, which is based on 'EUCAST Expert Rules' and 'EUCAST Intrinsic Resistance and Unusual Phenotypes' v3.3 (2021). The mo_is_intrinsic_resistant() functions can be vectorised over arguments x (input for microorganisms) and over ab (input for antibiotics).
All output will be translated where possible.
The function mo_url() will return the direct URL to the online database entry, which also shows the scientific reference of the concerned species.
SNOMED codes - mo_snomed() - are from the US Edition of SNOMED CT from 1 September 2020. See Source and the microorganisms data set for more info.
Matching Score for Microorganisms
With ambiguous user input in as.mo() and all the mo_* functions, the returned results are chosen based on their matching score using mo_matching_score(). This matching score \(m\), is calculated as:
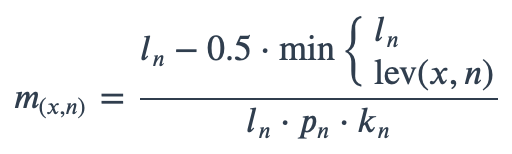
where:
x is the user input;
n is a taxonomic name (genus, species, and subspecies);
ln is the length of n;
lev is the Levenshtein distance function, which counts any insertion, deletion and substitution as 1 that is needed to change x into n;
pn is the human pathogenic prevalence group of n, as described below;
kn is the taxonomic kingdom of n, set as Bacteria = 1, Fungi = 2, Protozoa = 3, Archaea = 4, others = 5.
The grouping into human pathogenic prevalence (\(p\)) is based on experience from several microbiological laboratories in the Netherlands in conjunction with international reports on pathogen prevalence. Group 1 (most prevalent microorganisms) consists of all microorganisms where the taxonomic class is Gammaproteobacteria or where the taxonomic genus is Enterococcus, Staphylococcus or Streptococcus. This group consequently contains all common Gram-negative bacteria, such as Pseudomonas and Legionella and all species within the order Enterobacterales. Group 2 consists of all microorganisms where the taxonomic phylum is Proteobacteria, Firmicutes, Actinobacteria or Sarcomastigophora, or where the taxonomic genus is Absidia, Acremonium, Actinotignum, Alternaria, Anaerosalibacter, Apophysomyces, Arachnia, Aspergillus, Aureobacterium, Aureobasidium, Bacteroides, Basidiobolus, Beauveria, Blastocystis, Branhamella, Calymmatobacterium, Candida, Capnocytophaga, Catabacter, Chaetomium, Chryseobacterium, Chryseomonas, Chrysonilia, Cladophialophora, Cladosporium, Conidiobolus, Cryptococcus, Curvularia, Exophiala, Exserohilum, Flavobacterium, Fonsecaea, Fusarium, Fusobacterium, Hendersonula, Hypomyces, Koserella, Lelliottia, Leptosphaeria, Leptotrichia, Malassezia, Malbranchea, Mortierella, Mucor, Mycocentrospora, Mycoplasma, Nectria, Ochroconis, Oidiodendron, Phoma, Piedraia, Pithomyces, Pityrosporum, Prevotella, Pseudallescheria, Rhizomucor, Rhizopus, Rhodotorula, Scolecobasidium, Scopulariopsis, Scytalidium, Sporobolomyces, Stachybotrys, Stomatococcus, Treponema, Trichoderma, Trichophyton, Trichosporon, Tritirachium or Ureaplasma. Group 3 consists of all other microorganisms.
All characters in \(x\) and \(n\) are ignored that are other than A-Z, a-z, 0-9, spaces and parentheses.
All matches are sorted descending on their matching score and for all user input values, the top match will be returned. This will lead to the effect that e.g., "E. coli" will return the microbial ID of Escherichia coli (\(m = 0.688\), a highly prevalent microorganism found in humans) and not Entamoeba coli (\(m = 0.079\), a less prevalent microorganism in humans), although the latter would alphabetically come first.
Since AMR version 1.8.1, common microorganism abbreviations are ignored in determining the matching score. These abbreviations are currently: AIEC, ATEC, BORSA, CRSM, DAEC, EAEC, EHEC, EIEC, EPEC, ETEC, GISA, MRPA, MRSA, MRSE, MSSA, MSSE, NMEC, PISP, PRSP, STEC, UPEC, VISA, VISP, VRE, VRSA and VRSP.
Catalogue of Life
![]()
This package contains the complete taxonomic tree of almost all microorganisms (~71,000 species) from the authoritative and comprehensive Catalogue of Life (CoL, http://www.catalogueoflife.org). The CoL is the most comprehensive and authoritative global index of species currently available. Nonetheless, we supplemented the CoL data with data from the List of Prokaryotic names with Standing in Nomenclature (LPSN, lpsn.dsmz.de). This supplementation is needed until the CoL+ project is finished, which we await.
Click here for more information about the included taxa. Check which versions of the CoL and LPSN were included in this package with catalogue_of_life_version().
Source
Becker K et al. Coagulase-Negative Staphylococci. 2014. Clin Microbiol Rev. 27(4): 870-926; doi:10.1128/CMR.00109-13
Becker K et al. Implications of identifying the recently defined members of the S. aureus complex, S. argenteus and S. schweitzeri: A position paper of members of the ESCMID Study Group for staphylococci and Staphylococcal Diseases (ESGS). 2019. Clin Microbiol Infect; doi:10.1016/j.cmi.2019.02.028
Becker K et al. Emergence of coagulase-negative staphylococci 2020. Expert Rev Anti Infect Ther. 18(4):349-366; doi:10.1080/14787210.2020.1730813
Lancefield RC A serological differentiation of human and other groups of hemolytic streptococci. 1933. J Exp Med. 57(4): 571-95; doi:10.1084/jem.57.4.571
Catalogue of Life: 2019 Annual Checklist, http://www.catalogueoflife.org
List of Prokaryotic names with Standing in Nomenclature (5 October 2021), doi:10.1099/ijsem.0.004332
US Edition of SNOMED CT from 1 September 2020, retrieved from the Public Health Information Network Vocabulary Access and Distribution System (PHIN VADS), OID 2.16.840.1.114222.4.11.1009, version 12; url: https://phinvads.cdc.gov/vads/ViewValueSet.action?oid=2.16.840.1.114222.4.11.1009
Reference Data Publicly Available
All reference data sets (about microorganisms, antibiotics, R/SI interpretation, EUCAST rules, etc.) in this AMR package are publicly and freely available. We continually export our data sets to formats for use in R, SPSS, SAS, Stata and Excel. We also supply flat files that are machine-readable and suitable for input in any software program, such as laboratory information systems. Please find all download links on our website, which is automatically updated with every code change.
See also
Data set microorganisms
Examples
# taxonomic tree -----------------------------------------------------------
mo_kingdom("Klebsiella pneumoniae")
#> [1] "Bacteria"
mo_phylum("Klebsiella pneumoniae")
#> [1] "Proteobacteria"
mo_class("Klebsiella pneumoniae")
#> [1] "Gammaproteobacteria"
mo_order("Klebsiella pneumoniae")
#> [1] "Enterobacterales"
mo_family("Klebsiella pneumoniae")
#> [1] "Enterobacteriaceae"
mo_genus("Klebsiella pneumoniae")
#> [1] "Klebsiella"
mo_species("Klebsiella pneumoniae")
#> [1] "pneumoniae"
mo_subspecies("Klebsiella pneumoniae")
#> [1] ""
# colloquial properties ----------------------------------------------------
mo_name("Klebsiella pneumoniae")
#> [1] "Klebsiella pneumoniae"
mo_fullname("Klebsiella pneumoniae")
#> [1] "Klebsiella pneumoniae"
mo_shortname("Klebsiella pneumoniae")
#> [1] "K. pneumoniae"
# other properties ---------------------------------------------------------
mo_gramstain("Klebsiella pneumoniae")
#> [1] "Gram-negative"
mo_snomed("Klebsiella pneumoniae")
#> [1] 1.098101e+15 4.468700e+08 1.098201e+15 4.098010e+08 5.641501e+07
#> [6] 7.143150e+08
mo_type("Klebsiella pneumoniae")
#> [1] "Bacteria"
mo_rank("Klebsiella pneumoniae")
#> [1] "species"
mo_url("Klebsiella pneumoniae")
#> Klebsiella pneumoniae
#> "https://lpsn.dsmz.de/species/klebsiella-pneumoniae"
mo_synonyms("Klebsiella pneumoniae")
#> NULL
# scientific reference -----------------------------------------------------
mo_ref("Klebsiella pneumoniae")
#> [1] "Trevisan, 1887"
mo_authors("Klebsiella pneumoniae")
#> [1] "Trevisan"
mo_year("Klebsiella pneumoniae")
#> [1] 1887
mo_lpsn("Klebsiella pneumoniae")
#> [1] 777151
# abbreviations known in the field -----------------------------------------
mo_genus("MRSA")
#> [1] "Staphylococcus"
mo_species("MRSA")
#> [1] "aureus"
mo_shortname("VISA")
#> [1] "S. aureus"
mo_gramstain("VISA")
#> [1] "Gram-positive"
mo_genus("EHEC")
#> [1] "Escherichia"
mo_species("EHEC")
#> [1] "coli"
# known subspecies ---------------------------------------------------------
mo_name("doylei")
#> [1] "Campylobacter jejuni doylei"
mo_genus("doylei")
#> [1] "Campylobacter"
mo_species("doylei")
#> [1] "jejuni"
mo_subspecies("doylei")
#> [1] "doylei"
mo_fullname("K. pneu rh")
#> [1] "Klebsiella pneumoniae rhinoscleromatis"
mo_shortname("K. pneu rh")
#> [1] "K. pneumoniae"
# \donttest{
# Becker classification, see ?as.mo ----------------------------------------
mo_fullname("S. epi")
#> ℹ Function `as.mo()` is uncertain about "S. epi" (assuming Staphylococcus
#> epidermidis). Run `mo_uncertainties()` to review this.
#> [1] "Staphylococcus epidermidis"
mo_fullname("S. epi", Becker = TRUE)
#> ℹ Function `as.mo()` is uncertain about "S. epi" (assuming Staphylococcus
#> epidermidis). Run `mo_uncertainties()` to review this.
#> [1] "Coagulase-negative Staphylococcus (CoNS)"
mo_shortname("S. epi")
#> ℹ Function `as.mo()` is uncertain about "S. epi" (assuming Staphylococcus
#> epidermidis). Run `mo_uncertainties()` to review this.
#> [1] "S. epidermidis"
mo_shortname("S. epi", Becker = TRUE)
#> ℹ Function `as.mo()` is uncertain about "S. epi" (assuming Staphylococcus
#> epidermidis). Run `mo_uncertainties()` to review this.
#> [1] "CoNS"
# Lancefield classification, see ?as.mo ------------------------------------
mo_fullname("S. pyo")
#> [1] "Streptococcus pyogenes"
mo_fullname("S. pyo", Lancefield = TRUE)
#> [1] "Streptococcus group A"
mo_shortname("S. pyo")
#> [1] "S. pyogenes"
mo_shortname("S. pyo", Lancefield = TRUE)
#> [1] "GAS"
# language support --------------------------------------------------------
mo_gramstain("Klebsiella pneumoniae", language = "de")
#> [1] "Gramnegativ"
mo_gramstain("Klebsiella pneumoniae", language = "nl")
#> [1] "Gram-negatief"
mo_gramstain("Klebsiella pneumoniae", language = "es")
#> [1] "Gram negativo"
# mo_type is equal to mo_kingdom, but mo_kingdom will remain official
mo_kingdom("Klebsiella pneumoniae")
#> [1] "Bacteria"
mo_type("Klebsiella pneumoniae")
#> [1] "Bacteria"
mo_type("Klebsiella pneumoniae")
#> [1] "Bacteria"
mo_fullname("S. pyogenes",
Lancefield = TRUE,
language = "de")
#> [1] "Streptococcus Gruppe A"
mo_fullname("S. pyogenes",
Lancefield = TRUE,
language = "nl")
#> [1] "Streptococcus groep A"
# other --------------------------------------------------------------------
mo_is_yeast(c("Candida", "Trichophyton", "Klebsiella"))
#> [1] TRUE FALSE FALSE
# gram stains and intrinsic resistance can be used as a filter in dplyr verbs
if (require("dplyr")) {
example_isolates %>%
filter(mo_is_gram_positive())
example_isolates %>%
filter(mo_is_intrinsic_resistant(ab = "vanco"))
}
#> ℹ Using column 'mo' as input for `mo_is_gram_positive()`
#> ℹ Using column 'mo' as input for `mo_is_intrinsic_resistant()`
#> # A tibble: 709 × 49
#> date hospit…¹ ward_…² ward_…³ ward_…⁴ age gender patie…⁵ mo
#> <date> <fct> <lgl> <lgl> <lgl> <dbl> <chr> <chr> <mo>
#> 1 2002-01-02 D FALSE TRUE FALSE 65 F A77334 B_ESCHR_COLI
#> 2 2002-01-03 D FALSE TRUE FALSE 65 F A77334 B_ESCHR_COLI
#> 3 2002-01-19 D FALSE TRUE FALSE 71 M 738003 B_ESCHR_COLI
#> 4 2002-01-19 D FALSE TRUE FALSE 71 M 738003 B_ESCHR_COLI
#> 5 2002-01-21 D FALSE TRUE FALSE 75 F 462081 B_CTRBC_FRND
#> 6 2002-01-22 B TRUE FALSE FALSE 50 M F35553 B_PROTS_MRBL
#> 7 2002-01-22 B TRUE FALSE FALSE 50 M F35553 B_PROTS_MRBL
#> 8 2002-02-05 B TRUE FALSE FALSE 45 F 067927 B_SERRT_MRCS
#> 9 2002-02-05 B TRUE FALSE FALSE 45 F 067927 B_SERRT_MRCS
#> 10 2002-02-05 B TRUE FALSE FALSE 45 F 067927 B_SERRT_MRCS
#> # … with 699 more rows, 40 more variables: PEN <rsi>, OXA <rsi>, FLC <rsi>,
#> # AMX <rsi>, AMC <rsi>, AMP <rsi>, TZP <rsi>, CZO <rsi>, FEP <rsi>,
#> # CXM <rsi>, FOX <rsi>, CTX <rsi>, CAZ <rsi>, CRO <rsi>, GEN <rsi>,
#> # TOB <rsi>, AMK <rsi>, KAN <rsi>, TMP <rsi>, SXT <rsi>, NIT <rsi>,
#> # FOS <rsi>, LNZ <rsi>, CIP <rsi>, MFX <rsi>, VAN <rsi>, TEC <rsi>,
#> # TCY <rsi>, TGC <rsi>, DOX <rsi>, ERY <rsi>, CLI <rsi>, AZM <rsi>,
#> # IPM <rsi>, MEM <rsi>, MTR <rsi>, CHL <rsi>, COL <rsi>, MUP <rsi>, …
#> # ℹ Use `print(n = ...)` to see more rows, and `colnames()` to see all variable names
# get a list with the complete taxonomy (from kingdom to subspecies)
mo_taxonomy("Klebsiella pneumoniae")
#> $kingdom
#> [1] "Bacteria"
#>
#> $phylum
#> [1] "Proteobacteria"
#>
#> $class
#> [1] "Gammaproteobacteria"
#>
#> $order
#> [1] "Enterobacterales"
#>
#> $family
#> [1] "Enterobacteriaceae"
#>
#> $genus
#> [1] "Klebsiella"
#>
#> $species
#> [1] "pneumoniae"
#>
#> $subspecies
#> [1] ""
#>
# get a list with the taxonomy, the authors, Gram-stain,
# SNOMED codes, and URL to the online database
mo_info("Klebsiella pneumoniae")
#> $kingdom
#> [1] "Bacteria"
#>
#> $phylum
#> [1] "Proteobacteria"
#>
#> $class
#> [1] "Gammaproteobacteria"
#>
#> $order
#> [1] "Enterobacterales"
#>
#> $family
#> [1] "Enterobacteriaceae"
#>
#> $genus
#> [1] "Klebsiella"
#>
#> $species
#> [1] "pneumoniae"
#>
#> $subspecies
#> [1] ""
#>
#> $synonyms
#> NULL
#>
#> $gramstain
#> [1] "Gram-negative"
#>
#> $url
#> [1] "https://lpsn.dsmz.de/species/klebsiella-pneumoniae"
#>
#> $ref
#> [1] "Trevisan, 1887"
#>
#> $snomed
#> [1] 1.098101e+15 4.468700e+08 1.098201e+15 4.098010e+08 5.641501e+07
#> [6] 7.143150e+08
#>
# }