Benchmarks
Matthijs S. Berends
20 February 2019
benchmarks.RmdOne of the most important features of this package is the complete microbial taxonomic database, supplied by the Catalogue of Life (http://catalogueoflife.org). We created a function as.mo() that transforms any user input value to a valid microbial ID by using AI (Artificial Intelligence) combined with the taxonomic tree of Catalogue of Life.
Using the microbenchmark package, we can review the calculation performance of this function. Its function microbenchmark() runs different input expressions independently of each other and measures their time-to-result.
In the next test, we try to ‘coerce’ different input values for Staphylococcus aureus. The actual result is the same every time: it returns its MO code B_STPHY_AUR (B stands for Bacteria, the taxonomic kingdom).
But the calculation time differs a lot. Here, the AI effect can be reviewed best:
S.aureus <- microbenchmark(as.mo("sau"),
as.mo("stau"),
as.mo("staaur"),
as.mo("S. aureus"),
as.mo("S. aureus"),
as.mo("STAAUR"),
as.mo("Staphylococcus aureus"),
as.mo("B_STPHY_AUR"),
times = 10)
print(S.aureus, unit = "ms", signif = 3)
#> Unit: milliseconds
#> expr min lq mean median uq max
#> as.mo("sau") 42.500 42.800 44.200 43.100 43.900 53.700
#> as.mo("stau") 76.300 76.800 82.800 77.000 78.800 116.000
#> as.mo("staaur") 42.700 43.000 47.100 43.600 43.900 80.700
#> as.mo("S. aureus") 18.400 18.500 18.800 18.800 19.200 19.300
#> as.mo("S. aureus") 18.400 18.400 23.600 18.600 19.300 67.100
#> as.mo("STAAUR") 42.700 42.800 43.200 43.000 43.600 44.100
#> as.mo("Staphylococcus aureus") 11.400 11.500 11.700 11.600 11.800 12.500
#> as.mo("B_STPHY_AUR") 0.267 0.297 0.403 0.431 0.478 0.509
#> neval
#> 10
#> 10
#> 10
#> 10
#> 10
#> 10
#> 10
#> 10In the table above, all measurements are shown in milliseconds (thousands of seconds). A value of 10 milliseconds means it can determine 100 input values per second. It case of 50 milliseconds, this is only 20 input values per second. The more an input value resembles a full name, the faster the result will be found. In case of as.mo("B_STPHY_AUR"), the input is already a valid MO code, so it only almost takes no time at all (267 millionths of seconds).
To achieve this speed, the as.mo function also takes into account the prevalence of human pathogenic microorganisms. The downside is of course that less prevalent microorganisms will be determined less fast. See this example for the ID of Mycoplasma leonicaptivi (B_MYCPL_LEO), a bug probably never found before in humans:
M.leonicaptivi <- microbenchmark(as.mo("myle"),
as.mo("mycleo"),
as.mo("M. leonicaptivi"),
as.mo("M. leonicaptivi"),
as.mo("MYCLEO"),
as.mo("Mycoplasma leonicaptivi"),
as.mo("B_MYCPL_LEO"),
times = 10)
print(M.leonicaptivi, unit = "ms", signif = 4)
#> Unit: milliseconds
#> expr min lq mean median
#> as.mo("myle") 111.9000 112.0000 112.4000 112.2000
#> as.mo("mycleo") 381.4000 381.8000 388.5000 382.1000
#> as.mo("M. leonicaptivi") 203.0000 203.2000 212.5000 203.6000
#> as.mo("M. leonicaptivi") 203.0000 203.1000 212.7000 203.6000
#> as.mo("MYCLEO") 381.8000 382.4000 394.5000 382.9000
#> as.mo("Mycoplasma leonicaptivi") 102.8000 103.0000 103.4000 103.2000
#> as.mo("B_MYCPL_LEO") 0.3183 0.5657 0.5693 0.5727
#> uq max neval
#> 112.4000 113.5000 10
#> 385.4000 439.9000 10
#> 205.8000 253.9000 10
#> 207.2000 252.3000 10
#> 421.1000 422.1000 10
#> 103.4000 105.7000 10
#> 0.5994 0.7446 10That takes 6 times as much time on average! A value of 100 milliseconds means it can only determine ~10 different input values per second. We can conclude that looking up arbitrary codes of less prevalent microorganisms is the worst way to go, in terms of calculation performance:
par(mar = c(5, 16, 4, 2)) # set more space for left margin text (16)
# highest value on y axis
max_y_axis <- max(S.aureus$time, M.leonicaptivi$time, na.rm = TRUE) / 1e6
boxplot(S.aureus, horizontal = TRUE, las = 1, unit = "ms", log = FALSE, xlab = "", ylim = c(0, max_y_axis),
main = expression(paste("Benchmark of ", italic("Staphylococcus aureus"))))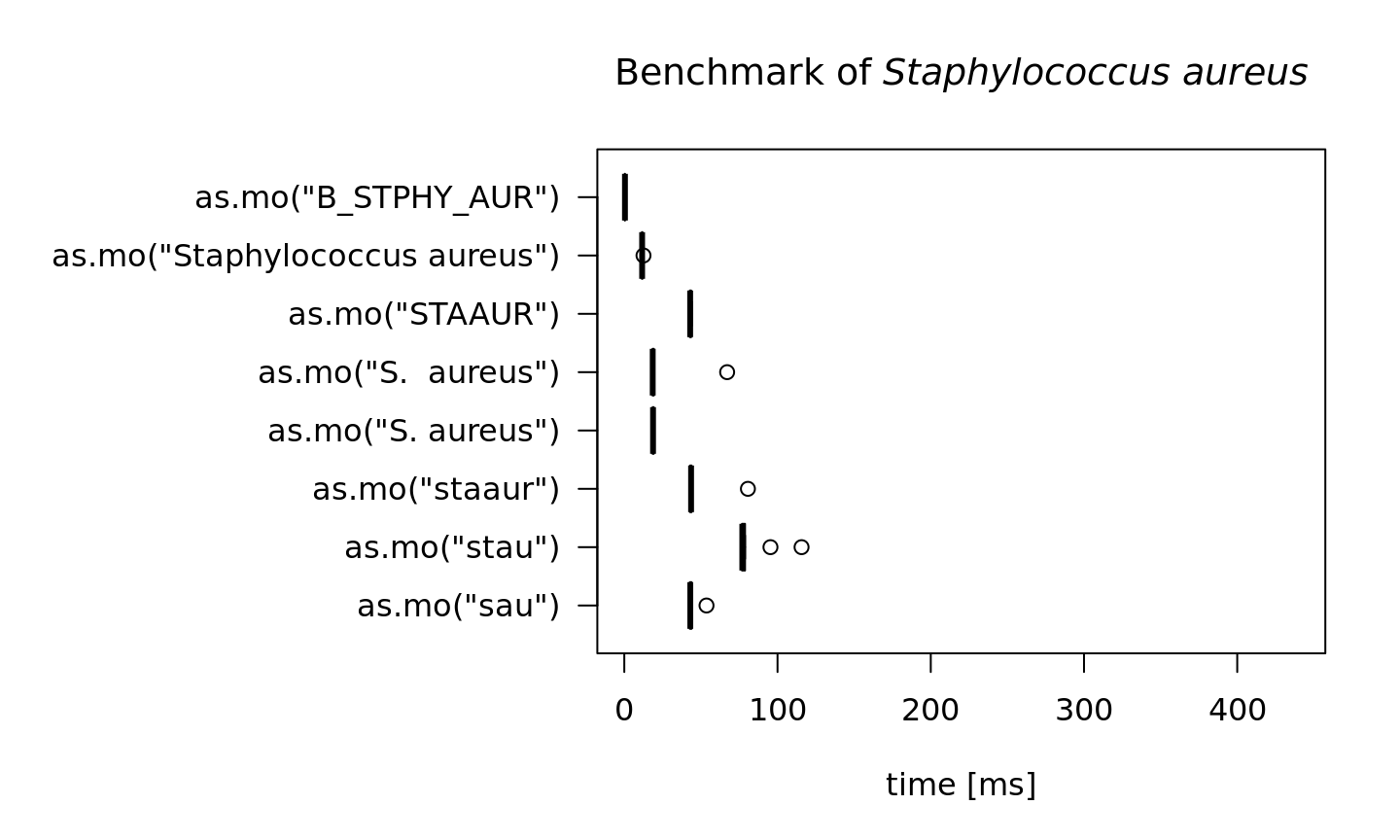
boxplot(M.leonicaptivi, horizontal = TRUE, las = 1, unit = "ms", log = FALSE, xlab = "", ylim = c(0, max_y_axis),
main = expression(paste("Benchmark of ", italic("Mycoplasma leonicaptivi"))))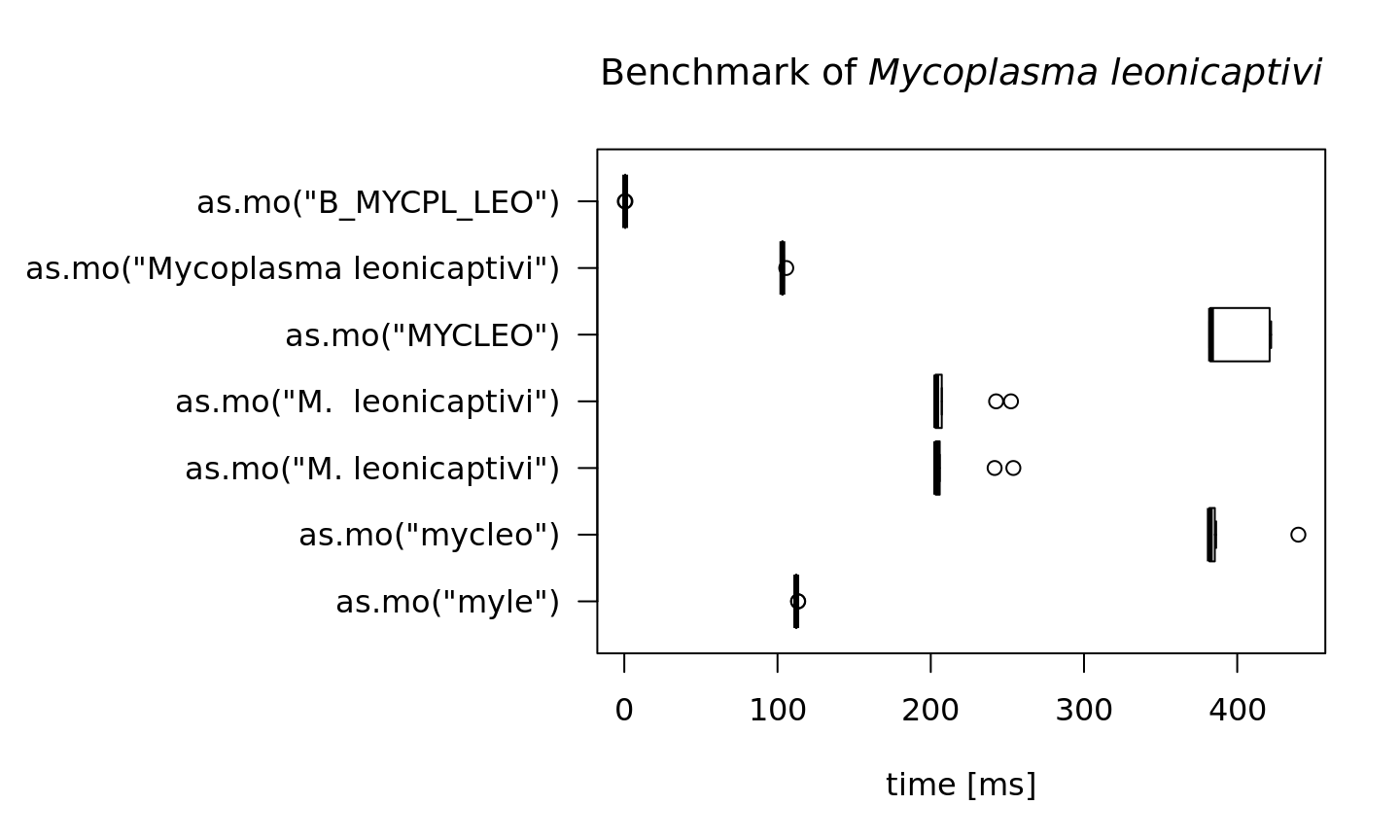
To relieve this pitfall and further improve performance, two important calculations take almost no time at all: repetitive results and already precalculated results.
Repetitive results
Repetitive results mean that unique values are present more than once. Unique values will only be calculated once by as.mo(). We will use mo_fullname() for this test - a helper function that returns the full microbial name (genus, species and possibly subspecies) which uses as.mo() internally.
library(dplyr)
# take 500,000 random MO codes from the septic_patients data set
x = septic_patients %>%
sample_n(500000, replace = TRUE) %>%
pull(mo)
# got the right length?
length(x)
#> [1] 500000
# and how many unique values do we have?
n_distinct(x)
#> [1] 95
# now let's see:
run_it <- microbenchmark(mo_fullname(x),
times = 10)
print(run_it, unit = "ms", signif = 3)
#> Unit: milliseconds
#> expr min lq mean median uq max neval
#> mo_fullname(x) 438 453 468 468 482 499 10So transforming 500,000 values (!) of 95 unique values only takes 0.47 seconds (468 ms). You only lose time on your unique input values.
Precalculated results
What about precalculated results? If the input is an already precalculated result of a helper function like mo_fullname(), it almost doesn’t take any time at all (see ‘C’ below):
run_it <- microbenchmark(A = mo_fullname("B_STPHY_AUR"),
B = mo_fullname("S. aureus"),
C = mo_fullname("Staphylococcus aureus"),
times = 10)
print(run_it, unit = "ms", signif = 3)
#> Unit: milliseconds
#> expr min lq mean median uq max neval
#> A 38.600 38.700 39.40 39.100 39.400 42.900 10
#> B 19.600 19.800 20.00 19.900 20.000 20.700 10
#> C 0.255 0.261 0.37 0.386 0.499 0.505 10So going from mo_fullname("Staphylococcus aureus") to "Staphylococcus aureus" takes 0.0004 seconds - it doesn’t even start calculating if the result would be the same as the expected resulting value. That goes for all helper functions:
run_it <- microbenchmark(A = mo_species("aureus"),
B = mo_genus("Staphylococcus"),
C = mo_fullname("Staphylococcus aureus"),
D = mo_family("Staphylococcaceae"),
E = mo_order("Bacillales"),
F = mo_class("Bacilli"),
G = mo_phylum("Firmicutes"),
H = mo_kingdom("Bacteria"),
times = 10)
print(run_it, unit = "ms", signif = 3)
#> Unit: milliseconds
#> expr min lq mean median uq max neval
#> A 0.282 0.311 0.372 0.359 0.437 0.513 10
#> B 0.285 0.316 0.355 0.363 0.382 0.443 10
#> C 0.258 0.408 0.439 0.430 0.504 0.565 10
#> D 0.268 0.304 0.322 0.321 0.360 0.366 10
#> E 0.259 0.273 0.312 0.295 0.357 0.391 10
#> F 0.250 0.275 0.327 0.294 0.343 0.614 10
#> G 0.254 0.281 0.312 0.320 0.338 0.369 10
#> H 0.257 0.265 0.311 0.316 0.329 0.397 10Of course, when running mo_phylum("Firmicutes") the function has zero knowledge about the actual microorganism, namely S. aureus. But since the result would be "Firmicutes" too, there is no point in calculating the result. And because this package ‘knows’ all phyla of all known bacteria (according to the Catalogue of Life), it can just return the initial value immediately.
Results in other languages
When the system language is non-English and supported by this AMR package, some functions will have a translated result. This almost does’t take extra time:
mo_fullname("CoNS", language = "en") # or just mo_fullname("CoNS") on an English system
#> [1] "Coagulase Negative Staphylococcus (CoNS)"
mo_fullname("CoNS", language = "es") # or just mo_fullname("CoNS") on a Spanish system
#> [1] "Staphylococcus coagulasa negativo (CoNS)"
mo_fullname("CoNS", language = "nl") # or just mo_fullname("CoNS") on a Dutch system
#> [1] "Coagulase-negatieve Staphylococcus (CNS)"
run_it <- microbenchmark(en = mo_fullname("CoNS", language = "en"),
de = mo_fullname("CoNS", language = "de"),
nl = mo_fullname("CoNS", language = "nl"),
es = mo_fullname("CoNS", language = "es"),
it = mo_fullname("CoNS", language = "it"),
fr = mo_fullname("CoNS", language = "fr"),
pt = mo_fullname("CoNS", language = "pt"),
times = 10)
print(run_it, unit = "ms", signif = 4)
#> Unit: milliseconds
#> expr min lq mean median uq max neval
#> en 10.69 11.02 11.06 11.08 11.21 11.29 10
#> de 19.09 19.48 19.49 19.55 19.62 19.66 10
#> nl 19.25 19.58 19.66 19.60 19.72 20.61 10
#> es 19.17 19.54 26.16 19.61 20.23 52.49 10
#> it 19.10 19.46 26.14 19.69 19.92 52.56 10
#> fr 19.10 19.36 19.50 19.50 19.59 20.13 10
#> pt 19.26 19.50 23.19 19.71 20.48 53.09 10Currently supported are German, Dutch, Spanish, Italian, French and Portuguese.