Benchmarks
Matthijs S. Berends
20 February 2019
benchmarks.RmdOne of the most important features of this package is the complete microbial taxonomic database, supplied by the Catalogue of Life (http://catalogueoflife.org). We created a function as.mo() that transforms any user input value to a valid microbial ID by using AI (Artificial Intelligence) combined with the taxonomic tree of Catalogue of Life.
Using the microbenchmark package, we can review the calculation performance of this function. Its function microbenchmark() runs different input expressions independently of each other and measures their time-to-result.
In the next test, we try to ‘coerce’ different input values for Staphylococcus aureus. The actual result is the same every time: it returns its MO code B_STPHY_AUR (B stands for Bacteria, the taxonomic kingdom).
But the calculation time differs a lot. Here, the AI effect can be reviewed best:
S.aureus <- microbenchmark(as.mo("sau"),
as.mo("stau"),
as.mo("staaur"),
as.mo("S. aureus"),
as.mo("S. aureus"),
as.mo("STAAUR"),
as.mo("Staphylococcus aureus"),
as.mo("B_STPHY_AUR"),
times = 10)
print(S.aureus, unit = "ms")
#> Unit: milliseconds
#> expr min lq mean median
#> as.mo("sau") 42.58139 42.645368 43.3006677 42.970095
#> as.mo("stau") 76.60094 77.168264 83.7686909 77.316642
#> as.mo("staaur") 42.86607 42.947083 43.5035571 43.497293
#> as.mo("S. aureus") 18.39354 18.432582 22.4304233 18.495928
#> as.mo("S. aureus") 18.46513 18.559903 18.6640991 18.579110
#> as.mo("STAAUR") 42.71975 42.788612 44.3280682 43.069864
#> as.mo("Staphylococcus aureus") 11.56285 11.591419 15.9457298 11.667161
#> as.mo("B_STPHY_AUR") 0.40487 0.450128 0.5036822 0.481417
#> uq max neval
#> 43.448543 45.058105 10
#> 78.335591 127.180349 10
#> 43.817095 44.999509 10
#> 19.007097 56.501460 10
#> 18.651814 19.373275 10
#> 43.741388 54.703256 10
#> 12.323077 50.121808 10
#> 0.519271 0.766578 10In the table above, all measurements are shown in milliseconds (thousands of seconds). A value of 10 milliseconds means it can determine 100 input values per second. It case of 50 milliseconds, this is only 20 input values per second. The more an input value resembles a full name, the faster the result will be found. In case of as.mo("B_STPHY_AUR"), the input is already a valid MO code, so it only almost takes no time at all (404 millionths of seconds).
To achieve this speed, the as.mo function also takes into account the prevalence of human pathogenic microorganisms. The downside is of course that less prevalent microorganisms will be determined far less faster. See this example for the ID of Mycoplasma leonicaptivi (B_MYCPL_LEO), a bug probably never found before in humans:
M.leonicaptivi <- microbenchmark(as.mo("myle"),
as.mo("mycleo"),
as.mo("M. leonicaptivi"),
as.mo("M. leonicaptivi"),
as.mo("MYCLEO"),
as.mo("Mycoplasma leonicaptivi"),
as.mo("B_MYCPL_LEO"),
times = 10)
print(M.leonicaptivi, unit = "ms")
#> Unit: milliseconds
#> expr min lq mean
#> as.mo("myle") 112.28656 112.372601 112.751678
#> as.mo("mycleo") 382.46812 382.757612 383.432440
#> as.mo("M. leonicaptivi") 202.68674 203.654949 210.461303
#> as.mo("M. leonicaptivi") 202.89759 203.440956 203.816387
#> as.mo("MYCLEO") 382.27864 383.090895 401.904482
#> as.mo("Mycoplasma leonicaptivi") 102.99676 103.191196 109.196394
#> as.mo("B_MYCPL_LEO") 0.32155 0.564807 4.320068
#> median uq max neval
#> 112.540884 112.76874 113.76321 10
#> 383.232219 384.05897 385.28587 10
#> 204.255445 205.80976 242.53035 10
#> 203.613673 203.82802 206.15038 10
#> 386.478757 421.87837 437.26978 10
#> 103.596136 104.65940 142.25748 10
#> 0.593652 0.62522 37.96384 10That takes 6 times as much time on average! A value of 100 milliseconds means it can only determine ~10 different input values per second. We can conclude that looking up arbitrary codes of less prevalent microorganisms is the worst way to go, in terms of calculation performance:
par(mar = c(5, 16, 4, 2)) # set more space for left margin text (16)
# highest value on y axis
max_y_axis <- max(S.aureus$time, M.leonicaptivi$time, na.rm = TRUE) / 1e6
boxplot(S.aureus, horizontal = TRUE, las = 1, unit = "ms", log = FALSE, xlab = "", ylim = c(0, max_y_axis),
main = expression(paste("Benchmark of ", italic("Staphylococcus aureus"))))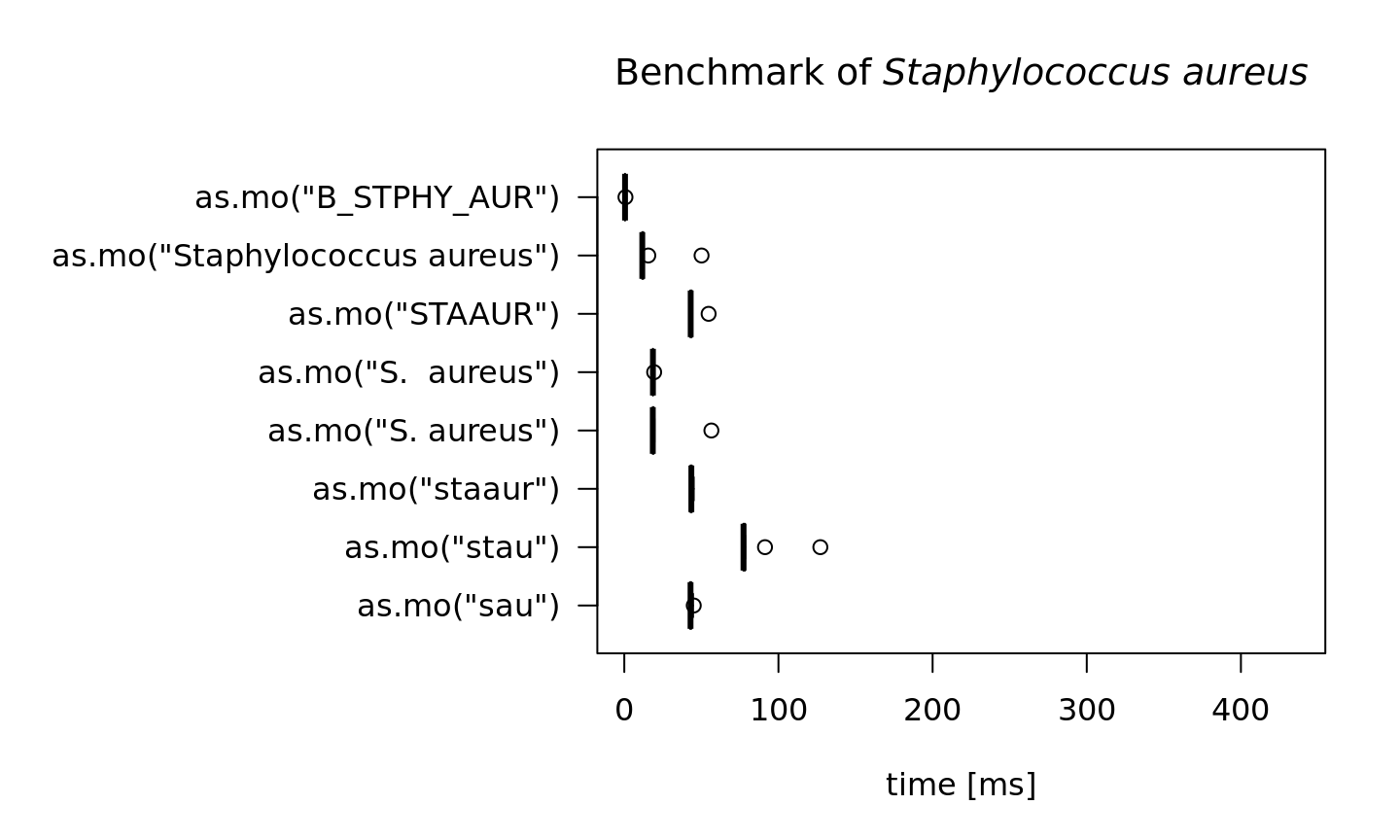
boxplot(M.leonicaptivi, horizontal = TRUE, las = 1, unit = "ms", log = FALSE, xlab = "", ylim = c(0, max_y_axis),
main = expression(paste("Benchmark of ", italic("Mycoplasma leonicaptivi"))))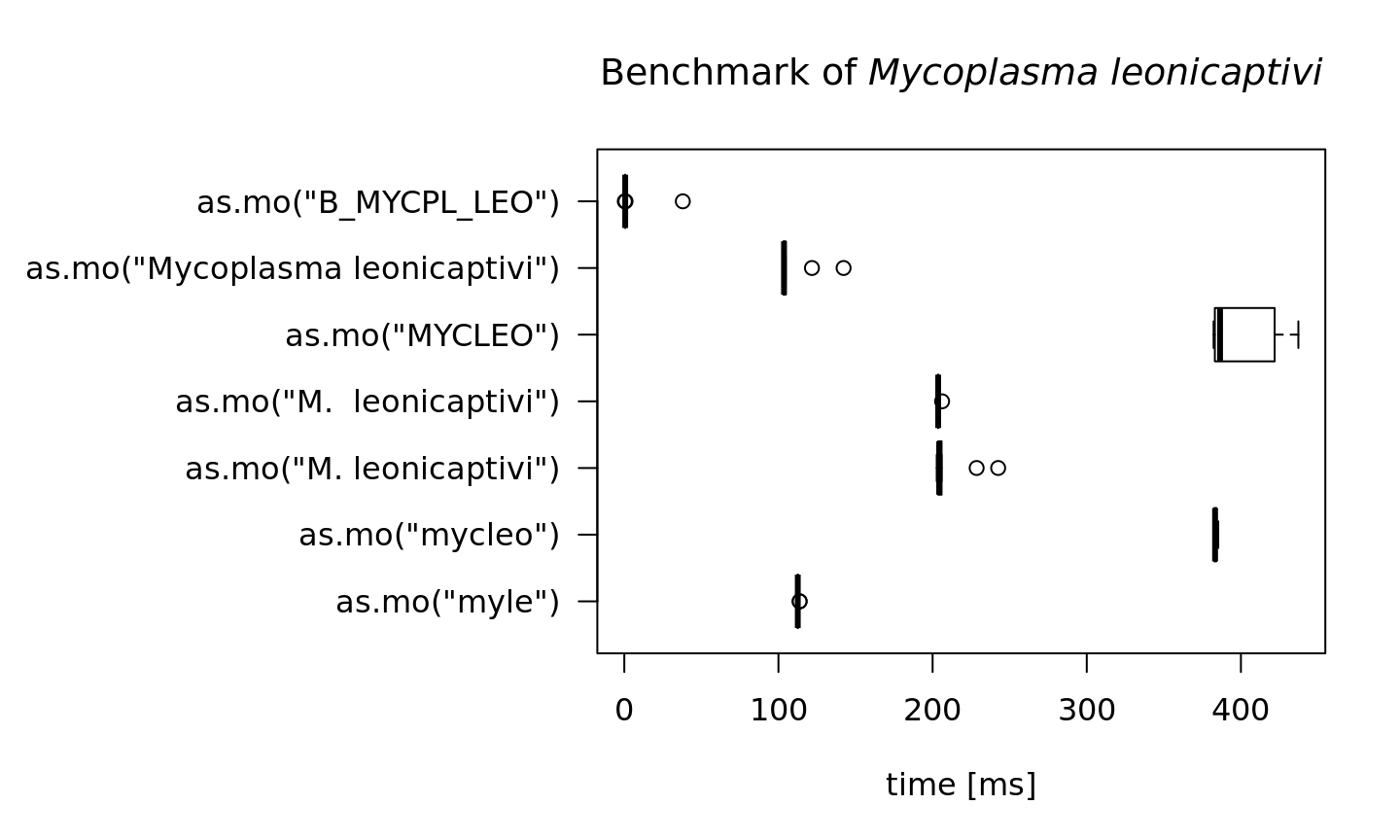
To relieve this pitfall and further improve performance, two important calculations take almost no time at all: repetitive results and already precalculated results.
Repetitive results
Repetitive results mean that unique values are present more than once. Unique values will only be calculated once by as.mo(). We will use mo_fullname() for this test - a helper function that returns the full microbial name (genus, species and possibly subspecies) which uses as.mo() internally.
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
# take 500,000 random MO codes from the septic_patients data set
x = septic_patients %>%
sample_n(500000, replace = TRUE) %>%
pull(mo)
# got the right length?
length(x)
#> [1] 500000
# and how many unique values do we have?
n_distinct(x)
#> [1] 95
# now let's see:
run_it <- microbenchmark(X = mo_fullname(x),
times = 10)
print(run_it, unit = "ms")
#> Unit: milliseconds
#> expr min lq mean median uq max neval
#> X 435.7086 442.1682 465.5949 468.8453 477.1915 505.961 10So transforming 500,000 values (!) of 95 unique values only takes 0.47 seconds (468 ms). You only lose time on your unique input values.
Precalculated results
What about precalculated results? If the input is an already precalculated result of a helper function like mo_fullname(), it almost doesn’t take any time at all (see ‘C’ below):
run_it <- microbenchmark(A = mo_fullname("B_STPHY_AUR"),
B = mo_fullname("S. aureus"),
C = mo_fullname("Staphylococcus aureus"),
times = 10)
print(run_it, unit = "ms")
#> Unit: milliseconds
#> expr min lq mean median uq max neval
#> A 38.887977 38.920313 39.3674024 39.076862 39.258415 42.166327 10
#> B 19.589084 19.631059 19.8682396 19.781567 19.955611 20.751941 10
#> C 0.255829 0.382732 0.4199913 0.400156 0.499156 0.564807 10So going from mo_fullname("Staphylococcus aureus") to "Staphylococcus aureus" takes 0.0004 seconds - it doesn’t even start calculating if the result would be the same as the expected resulting value. That goes for all helper functions:
microbenchmark(A = mo_species("aureus"),
B = mo_genus("Staphylococcus"),
C = mo_fullname("Staphylococcus aureus"),
D = mo_family("Staphylococcaceae"),
E = mo_order("Bacillales"),
F = mo_class("Bacilli"),
G = mo_phylum("Firmicutes"),
H = mo_kingdom("Bacteria"),
times = 10,
unit = "ms")
#> Unit: milliseconds
#> expr min lq mean median uq max neval
#> A 0.250242 0.292496 0.3891774 0.4266960 0.456902 0.520388 10
#> B 0.259461 0.311702 0.3428727 0.3412800 0.374141 0.443912 10
#> C 0.290960 0.313169 0.4334429 0.4097595 0.520389 0.725373 10
#> D 0.271823 0.282789 0.3187217 0.3192800 0.352909 0.375398 10
#> E 0.245353 0.270985 0.3081197 0.2960235 0.330839 0.429036 10
#> F 0.246122 0.266585 0.2991101 0.3089435 0.332794 0.351582 10
#> G 0.271893 0.272452 0.3085039 0.2850580 0.368204 0.385525 10
#> H 0.252686 0.259251 0.3161791 0.2985025 0.334820 0.422680 10Of course, when running mo_phylum("Firmicutes") the function has zero knowledge about the actual microorganism, namely S. aureus. But since the result would be "Firmicutes" too, there is no point in calculating the result. And because this package ‘knows’ all phyla of all known bacteria (according to the Catalogue of Life), it can just return the initial value immediately.
Results in other languages
When the system language is non-English and supported by this AMR package, some functions take a little while longer:
mo_fullname("CoNS", language = "en") # or just mo_fullname("CoNS") on an English system
#> [1] "Coagulase Negative Staphylococcus (CoNS)"
mo_fullname("CoNS", language = "fr") # or just mo_fullname("CoNS") on a French system
#> [1] "Staphylococcus à coagulase négative (CoNS)"
microbenchmark(en = mo_fullname("CoNS", language = "en"),
de = mo_fullname("CoNS", language = "de"),
nl = mo_fullname("CoNS", language = "nl"),
es = mo_fullname("CoNS", language = "es"),
it = mo_fullname("CoNS", language = "it"),
fr = mo_fullname("CoNS", language = "fr"),
pt = mo_fullname("CoNS", language = "pt"),
times = 10,
unit = "ms")
#> Unit: milliseconds
#> expr min lq mean median uq max neval
#> en 10.67105 11.03136 11.06332 11.07271 11.15310 11.45006 10
#> de 19.13393 19.50080 26.13799 19.61419 20.23400 52.66501 10
#> nl 19.05410 19.53789 22.94707 19.59205 20.12616 52.47399 10
#> es 19.31635 19.55221 26.22342 19.58633 20.01875 52.97636 10
#> it 19.21725 19.47105 19.63980 19.58053 19.68162 20.58914 10
#> fr 19.07854 19.45450 19.67303 19.56153 19.64517 20.45651 10
#> pt 19.00668 19.28388 19.53493 19.57857 19.66423 20.55317 10Currently supported are German, Dutch, Spanish, Italian, French and Portuguese.