Benchmarks
Matthijs S. Berends
20 February 2019
benchmarks.RmdOne of the most important features of this package is the complete microbial taxonomic database, supplied by the Catalogue of Life (http://catalogueoflife.org). We created a function as.mo() that transforms any user input value to a valid microbial ID by using AI (Artificial Intelligence) combined with the taxonomic tree of Catalogue of Life.
Using the microbenchmark package, we can review the calculation performance of this function. Its function microbenchmark() runs different input expressions independently of each other and measures their time-to-result.
In the next test, we try to ‘coerce’ different input values for Staphylococcus aureus. The actual result is the same every time: it returns its MO code B_STPHY_AUR (B stands for Bacteria, the taxonomic kingdom).
But the calculation time differs a lot. Here, the AI effect can be reviewed best:
S.aureus <- microbenchmark(as.mo("sau"),
as.mo("stau"),
as.mo("staaur"),
as.mo("S. aureus"),
as.mo("S. aureus"),
as.mo("STAAUR"),
as.mo("Staphylococcus aureus"),
as.mo("B_STPHY_AUR"),
times = 10)
print(S.aureus, unit = "ms")
#> Unit: milliseconds
#> expr min lq mean median
#> as.mo("sau") 42.680497 42.766053 43.5046242 43.2246305
#> as.mo("stau") 76.627901 76.760320 82.2084011 77.2020310
#> as.mo("staaur") 42.751945 42.828281 46.8816599 43.0017665
#> as.mo("S. aureus") 18.328588 18.370632 22.3298018 18.4252830
#> as.mo("S. aureus") 18.258048 18.385997 18.7710600 18.5449555
#> as.mo("STAAUR") 42.734554 42.854751 43.6593017 43.6353320
#> as.mo("Staphylococcus aureus") 11.466961 11.572841 16.5287637 11.6172940
#> as.mo("B_STPHY_AUR") 0.284603 0.302692 0.4095492 0.4190475
#> uq max neval
#> 44.091919 45.191431 10
#> 78.670409 123.715942 10
#> 43.089558 81.640969 10
#> 18.546004 57.384741 10
#> 19.235128 19.693775 10
#> 44.189907 45.381609 10
#> 12.175081 59.815567 10
#> 0.482254 0.500343 10In the table above, all measurements are shown in milliseconds (thousands of seconds). A value of 10 milliseconds means it can determine 100 input values per second. It case of 50 milliseconds, this is only 20 input values per second. The more an input value resembles a full name, the faster the result will be found. In case of as.mo("B_STPHY_AUR"), the input is already a valid MO code, so it only almost takes no time at all (284 millionths of seconds).
To achieve this speed, the as.mo function also takes into account the prevalence of human pathogenic microorganisms. The downside is of course that less prevalent microorganisms will be determined less fast. See this example for the ID of Mycoplasma leonicaptivi (B_MYCPL_LEO), a bug probably never found before in humans:
M.leonicaptivi <- microbenchmark(as.mo("myle"),
as.mo("mycleo"),
as.mo("M. leonicaptivi"),
as.mo("M. leonicaptivi"),
as.mo("MYCLEO"),
as.mo("Mycoplasma leonicaptivi"),
as.mo("B_MYCPL_LEO"),
times = 10)
print(M.leonicaptivi, unit = "ms")
#> Unit: milliseconds
#> expr min lq mean
#> as.mo("myle") 112.493914 112.698409 113.5834588
#> as.mo("mycleo") 382.813554 382.992838 389.0918181
#> as.mo("M. leonicaptivi") 202.903596 203.855253 211.7932317
#> as.mo("M. leonicaptivi") 203.761037 204.178479 212.5451427
#> as.mo("MYCLEO") 382.602355 383.481517 393.2696052
#> as.mo("Mycoplasma leonicaptivi") 103.701176 103.991018 109.5707840
#> as.mo("B_MYCPL_LEO") 0.312051 0.564876 0.5870787
#> median uq max neval
#> 113.363438 114.20691 115.907686 10
#> 384.139806 388.34114 421.458483 10
#> 204.186195 205.16631 243.204461 10
#> 204.715173 207.97372 244.462163 10
#> 383.918409 386.97938 434.456156 10
#> 104.428888 104.87207 153.125617 10
#> 0.567914 0.63779 0.859048 10That takes 6 times as much time on average! A value of 100 milliseconds means it can only determine ~10 different input values per second. We can conclude that looking up arbitrary codes of less prevalent microorganisms is the worst way to go, in terms of calculation performance:
par(mar = c(5, 16, 4, 2)) # set more space for left margin text (16)
# highest value on y axis
max_y_axis <- max(S.aureus$time, M.leonicaptivi$time, na.rm = TRUE) / 1e6
boxplot(S.aureus, horizontal = TRUE, las = 1, unit = "ms", log = FALSE, xlab = "", ylim = c(0, max_y_axis),
main = expression(paste("Benchmark of ", italic("Staphylococcus aureus"))))
boxplot(M.leonicaptivi, horizontal = TRUE, las = 1, unit = "ms", log = FALSE, xlab = "", ylim = c(0, max_y_axis),
main = expression(paste("Benchmark of ", italic("Mycoplasma leonicaptivi"))))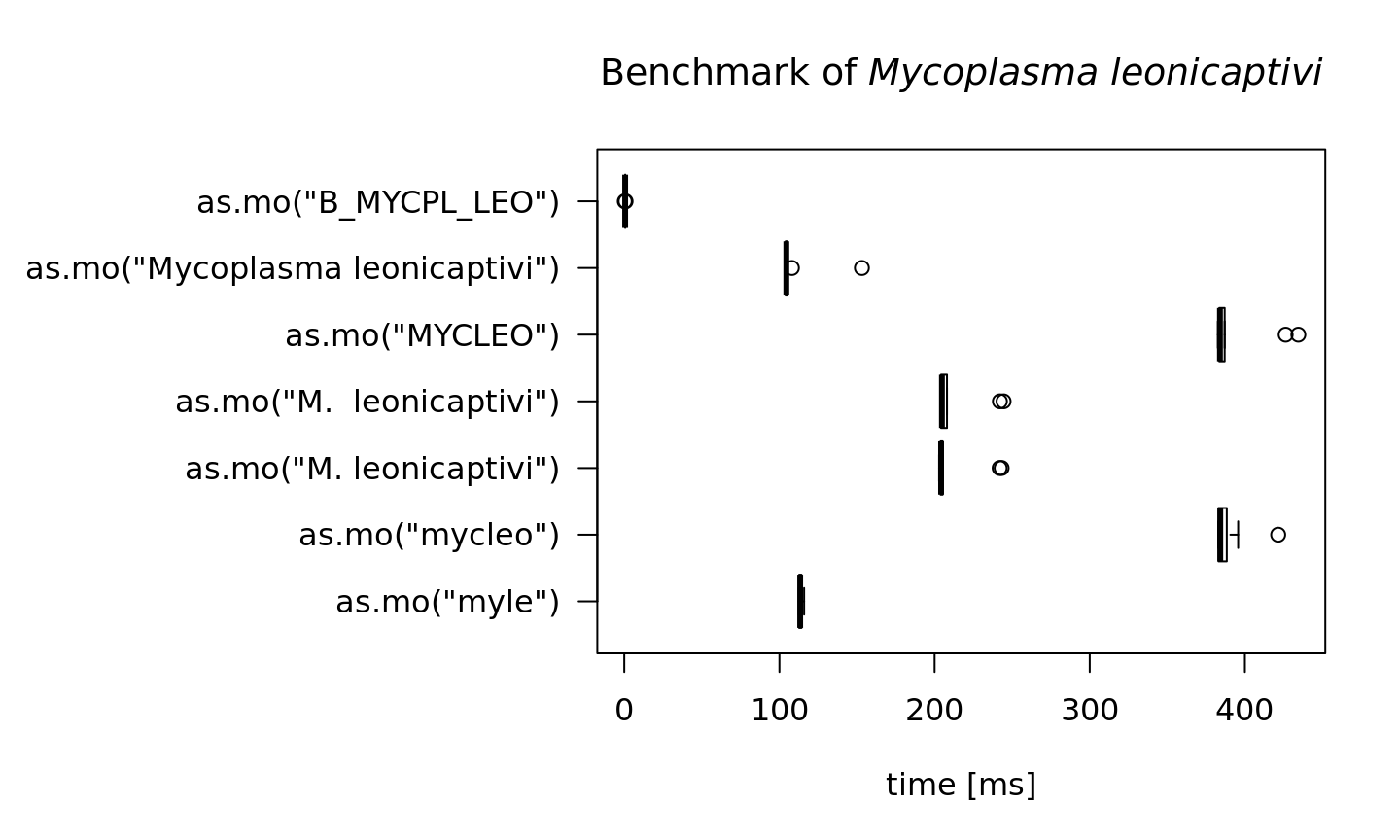
To relieve this pitfall and further improve performance, two important calculations take almost no time at all: repetitive results and already precalculated results.
Repetitive results
Repetitive results mean that unique values are present more than once. Unique values will only be calculated once by as.mo(). We will use mo_fullname() for this test - a helper function that returns the full microbial name (genus, species and possibly subspecies) which uses as.mo() internally.
library(dplyr)
# take 500,000 random MO codes from the septic_patients data set
x = septic_patients %>%
sample_n(500000, replace = TRUE) %>%
pull(mo)
# got the right length?
length(x)
#> [1] 500000
# and how many unique values do we have?
n_distinct(x)
#> [1] 95
# now let's see:
run_it <- microbenchmark(X = mo_fullname(x),
times = 10)
print(run_it, unit = "ms")
#> Unit: milliseconds
#> expr min lq mean median uq max neval
#> X 413.2556 431.8327 448.3355 445.8654 465.2447 480.5499 10So transforming 500,000 values (!) of 95 unique values only takes 0.45 seconds (445 ms). You only lose time on your unique input values.
Precalculated results
What about precalculated results? If the input is an already precalculated result of a helper function like mo_fullname(), it almost doesn’t take any time at all (see ‘C’ below):
run_it <- microbenchmark(A = mo_fullname("B_STPHY_AUR"),
B = mo_fullname("S. aureus"),
C = mo_fullname("Staphylococcus aureus"),
times = 10)
print(run_it, unit = "ms")
#> Unit: milliseconds
#> expr min lq mean median uq max neval
#> A 39.603291 39.713640 39.950479 39.8150500 40.172707 40.664181 10
#> B 19.570436 19.623515 19.964292 19.9376620 20.228830 20.609744 10
#> C 0.251429 0.333144 0.389883 0.3866775 0.499087 0.510401 10So going from mo_fullname("Staphylococcus aureus") to "Staphylococcus aureus" takes 0.0004 seconds - it doesn’t even start calculating if the result would be the same as the expected resulting value. That goes for all helper functions:
microbenchmark(A = mo_species("aureus"),
B = mo_genus("Staphylococcus"),
C = mo_fullname("Staphylococcus aureus"),
D = mo_family("Staphylococcaceae"),
E = mo_order("Bacillales"),
F = mo_class("Bacilli"),
G = mo_phylum("Firmicutes"),
H = mo_kingdom("Bacteria"),
times = 10,
unit = "ms")
#> Unit: milliseconds
#> expr min lq mean median uq max neval
#> A 0.298084 0.370509 0.4040816 0.4065820 0.449569 0.475480 10
#> B 0.293753 0.306115 0.3352809 0.3212705 0.370160 0.386154 10
#> C 0.307652 0.353328 0.4106327 0.3943595 0.467239 0.548255 10
#> D 0.244376 0.262954 0.2987189 0.3027975 0.338102 0.353747 10
#> E 0.249614 0.255550 0.2985027 0.2772710 0.351931 0.397049 10
#> F 0.259531 0.282439 0.3248814 0.3193850 0.345575 0.415906 10
#> G 0.249055 0.266516 0.3293723 0.3020295 0.344528 0.616350 10
#> H 0.242141 0.288515 0.3122614 0.3152295 0.339779 0.355773 10Of course, when running mo_phylum("Firmicutes") the function has zero knowledge about the actual microorganism, namely S. aureus. But since the result would be "Firmicutes" too, there is no point in calculating the result. And because this package ‘knows’ all phyla of all known bacteria (according to the Catalogue of Life), it can just return the initial value immediately.
Results in other languages
When the system language is non-English and supported by this AMR package, some functions will have a translated result. This almost does’t take extra time:
mo_fullname("CoNS", language = "en") # or just mo_fullname("CoNS") on an English system
#> [1] "Coagulase Negative Staphylococcus (CoNS)"
mo_fullname("CoNS", language = "fr") # or just mo_fullname("CoNS") on a French system
#> [1] "Staphylococcus à coagulase négative (CoNS)"
microbenchmark(en = mo_fullname("CoNS", language = "en"),
de = mo_fullname("CoNS", language = "de"),
nl = mo_fullname("CoNS", language = "nl"),
es = mo_fullname("CoNS", language = "es"),
it = mo_fullname("CoNS", language = "it"),
fr = mo_fullname("CoNS", language = "fr"),
pt = mo_fullname("CoNS", language = "pt"),
times = 10,
unit = "ms")
#> Unit: milliseconds
#> expr min lq mean median uq max neval
#> en 10.74026 11.10686 11.09997 11.11563 11.20366 11.34076 10
#> de 19.15977 19.59293 19.76980 19.71204 19.78338 20.54633 10
#> nl 19.42929 19.54013 19.75978 19.67233 19.77263 20.58935 10
#> es 19.31042 19.66821 19.65120 19.69552 19.73421 19.75538 10
#> it 19.26362 19.34003 22.93301 19.62998 19.67213 52.79729 10
#> fr 19.33011 19.54739 26.16391 19.64726 19.87145 52.40164 10
#> pt 19.22800 19.50164 26.41786 19.66766 20.96244 53.16479 10Currently supported are German, Dutch, Spanish, Italian, French and Portuguese.