Functions to plot classes sir, mic and disk, with support for base R and ggplot2.
Usage
# S3 method for mic
plot(
x,
mo = NULL,
ab = NULL,
guideline = "EUCAST",
main = deparse(substitute(x)),
ylab = "Frequency",
xlab = "Minimum Inhibitory Concentration (mg/L)",
colours_SIR = c("#3CAEA3", "#F6D55C", "#ED553B"),
language = get_AMR_locale(),
expand = TRUE,
...
)
# S3 method for mic
autoplot(
object,
mo = NULL,
ab = NULL,
guideline = "EUCAST",
title = deparse(substitute(object)),
ylab = "Frequency",
xlab = "Minimum Inhibitory Concentration (mg/L)",
colours_SIR = c("#3CAEA3", "#F6D55C", "#ED553B"),
language = get_AMR_locale(),
expand = TRUE,
...
)
# S3 method for mic
fortify(object, ...)
# S3 method for disk
plot(
x,
main = deparse(substitute(x)),
ylab = "Frequency",
xlab = "Disk diffusion diameter (mm)",
mo = NULL,
ab = NULL,
guideline = "EUCAST",
colours_SIR = c("#3CAEA3", "#F6D55C", "#ED553B"),
language = get_AMR_locale(),
expand = TRUE,
...
)
# S3 method for disk
autoplot(
object,
mo = NULL,
ab = NULL,
title = deparse(substitute(object)),
ylab = "Frequency",
xlab = "Disk diffusion diameter (mm)",
guideline = "EUCAST",
colours_SIR = c("#3CAEA3", "#F6D55C", "#ED553B"),
language = get_AMR_locale(),
expand = TRUE,
...
)
# S3 method for disk
fortify(object, ...)
# S3 method for sir
plot(
x,
ylab = "Percentage",
xlab = "Antimicrobial Interpretation",
main = deparse(substitute(x)),
language = get_AMR_locale(),
...
)
# S3 method for sir
autoplot(
object,
title = deparse(substitute(object)),
xlab = "Antimicrobial Interpretation",
ylab = "Frequency",
colours_SIR = c("#3CAEA3", "#F6D55C", "#ED553B"),
language = get_AMR_locale(),
...
)
# S3 method for sir
fortify(object, ...)Arguments
- x, object
values created with
as.mic(),as.disk()oras.sir()(or theirrandom_*variants, such asrandom_mic())- mo
any (vector of) text that can be coerced to a valid microorganism code with
as.mo()- ab
any (vector of) text that can be coerced to a valid antimicrobial drug code with
as.ab()- guideline
interpretation guideline to use, defaults to the latest included EUCAST guideline, see Details
- main, title
title of the plot
- xlab, ylab
axis title
- colours_SIR
colours to use for filling in the bars, must be a vector of three values (in the order S, I and R). The default colours are colour-blind friendly.
- language
language to be used to translate 'Susceptible', 'Increased exposure'/'Intermediate' and 'Resistant', defaults to system language (see
get_AMR_locale()) and can be overwritten by setting the optionAMR_locale, e.g.options(AMR_locale = "de"), see translate. Uselanguage = NULLorlanguage = ""to prevent translation.- expand
a logical to indicate whether the range on the x axis should be expanded between the lowest and highest value. For MIC values, intermediate values will be factors of 2 starting from the highest MIC value. For disk diameters, the whole diameter range will be filled.
- ...
arguments passed on to methods
Value
The autoplot() functions return a ggplot model that is extendible with any ggplot2 function.
The fortify() functions return a data.frame as an extension for usage in the ggplot2::ggplot() function.
Details
The interpretation of "I" will be named "Increased exposure" for all EUCAST guidelines since 2019, and will be named "Intermediate" in all other cases.
For interpreting MIC values as well as disk diffusion diameters, supported guidelines to be used as input for the guideline argument are: "EUCAST 2022", "EUCAST 2021", "EUCAST 2020", "EUCAST 2019", "EUCAST 2018", "EUCAST 2017", "EUCAST 2016", "EUCAST 2015", "EUCAST 2014", "EUCAST 2013", "CLSI 2022", "CLSI 2021", "CLSI 2020", "CLSI 2019", "CLSI 2018", "CLSI 2017", "CLSI 2016", "CLSI 2015", "CLSI 2014" and "CLSI 2013".
Simply using "CLSI" or "EUCAST" as input will automatically select the latest version of that guideline.
Examples
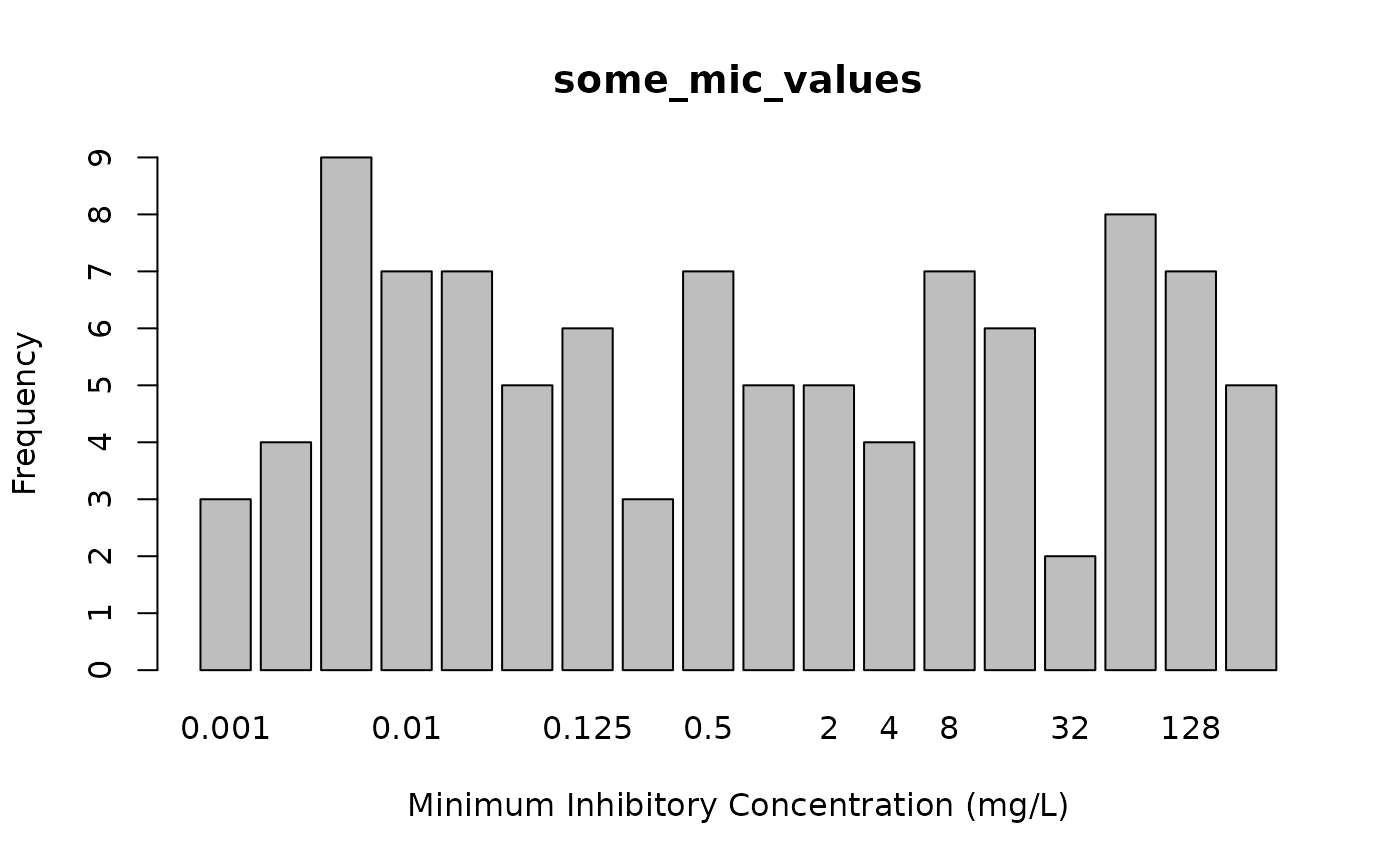
some_mic_values <- random_mic(size = 100)
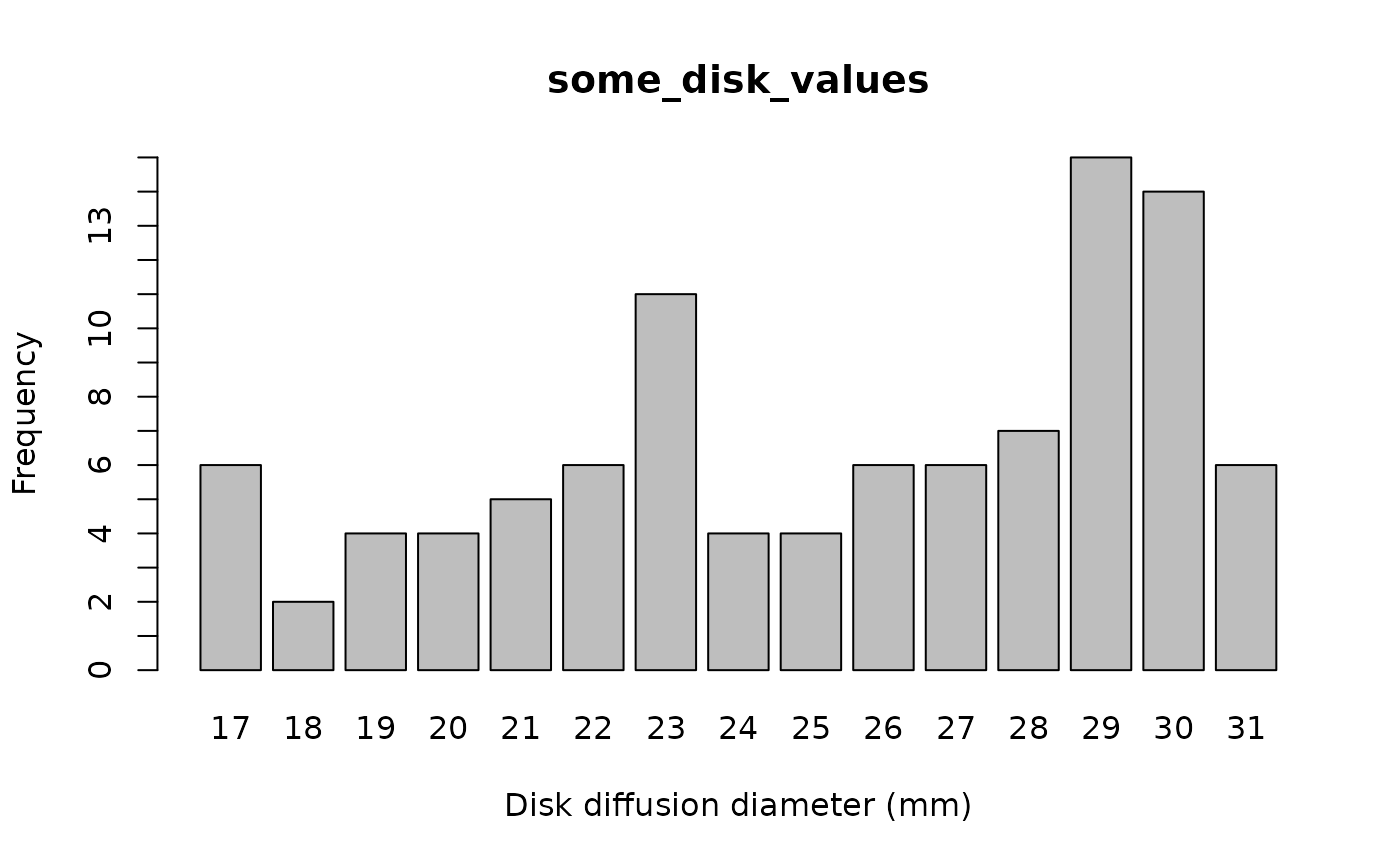
some_disk_values <- random_disk(size = 100, mo = "Escherichia coli", ab = "cipro")
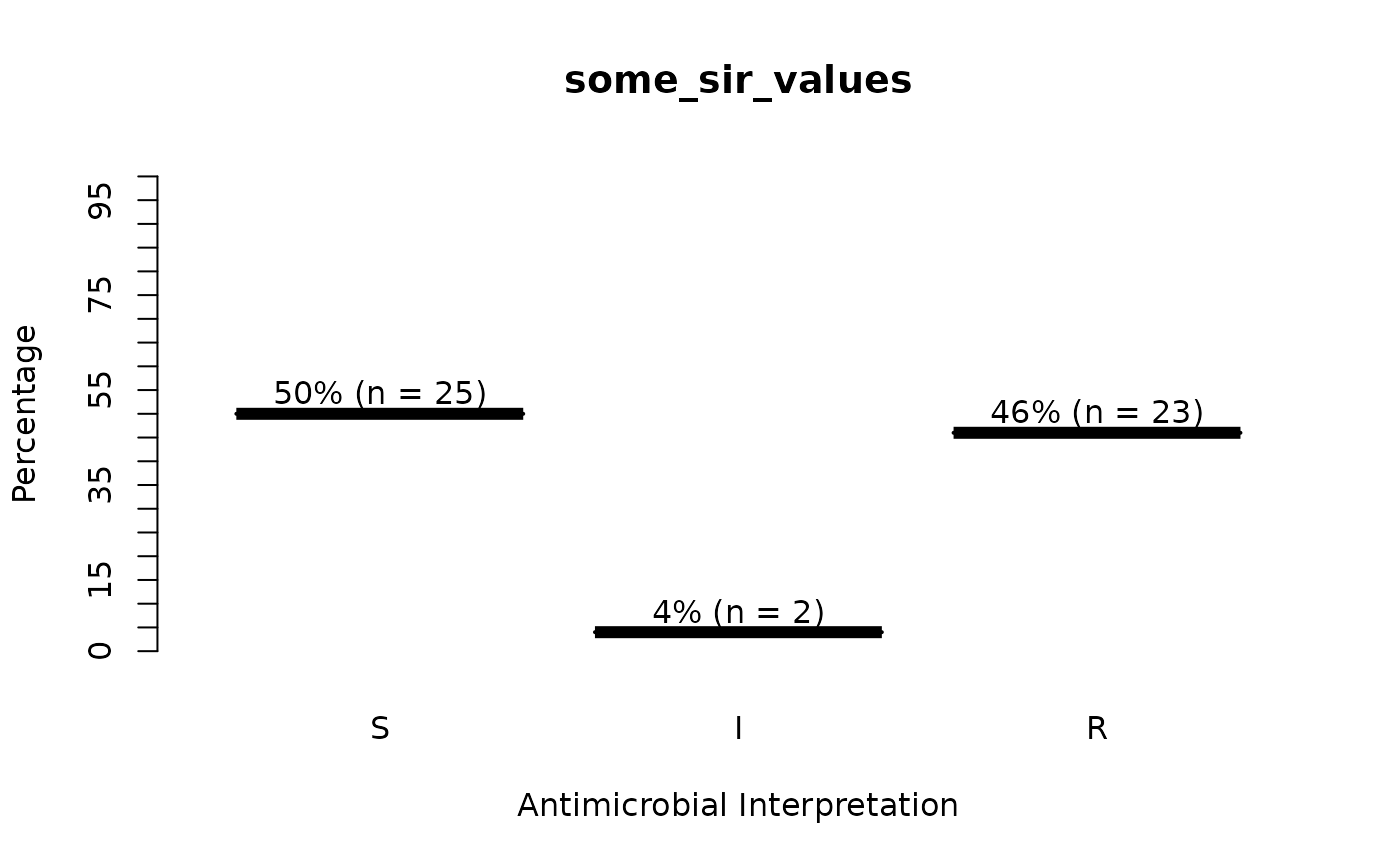
some_sir_values <- random_sir(50, prob_SIR = c(0.55, 0.05, 0.30))
plot(some_mic_values)
 plot(some_disk_values)
plot(some_disk_values)
 plot(some_sir_values)
plot(some_sir_values)
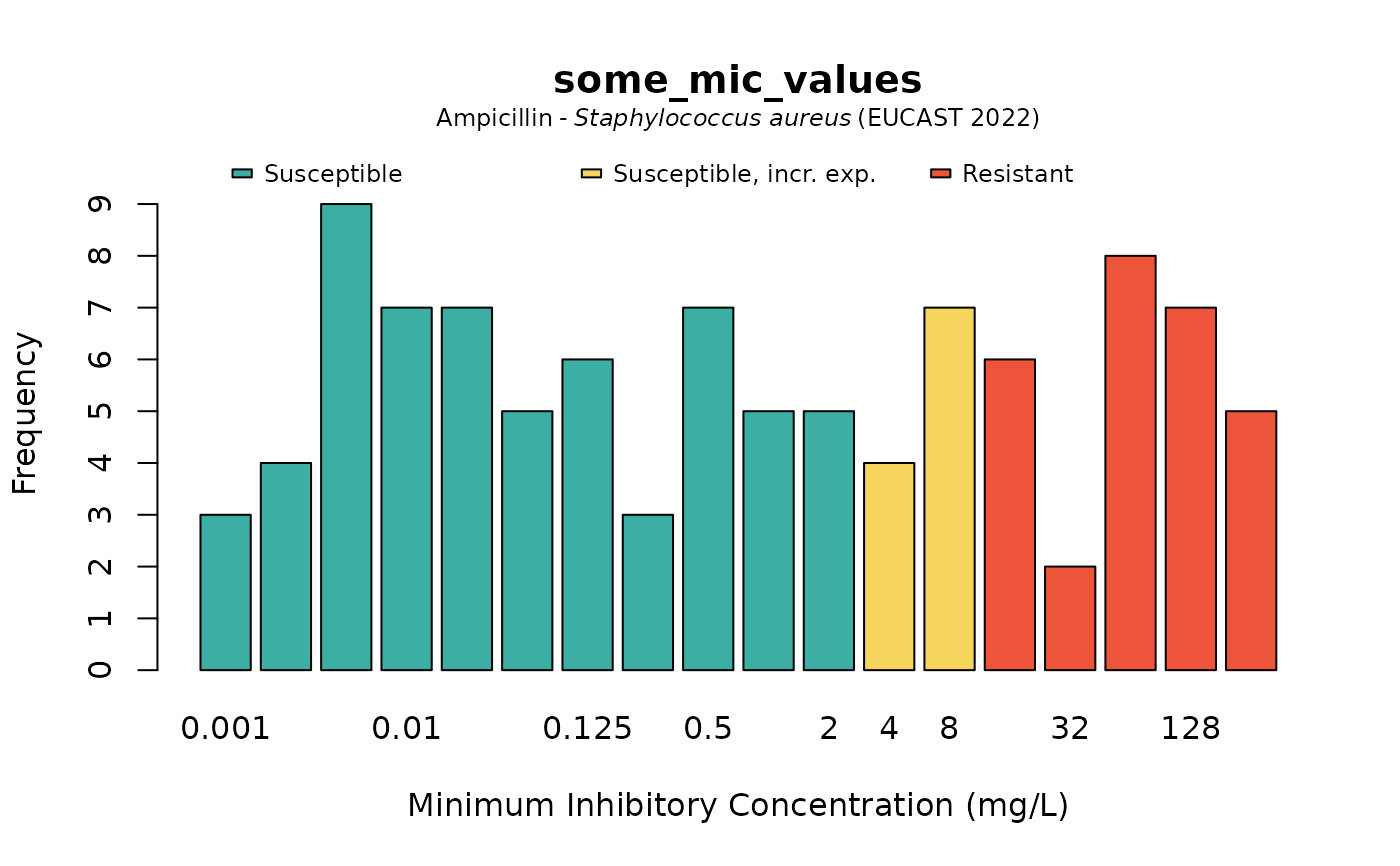
 # when providing the microorganism and antibiotic, colours will show interpretations:
plot(some_mic_values, mo = "S. aureus", ab = "ampicillin")
# when providing the microorganism and antibiotic, colours will show interpretations:
plot(some_mic_values, mo = "S. aureus", ab = "ampicillin")
 plot(some_disk_values, mo = "Escherichia coli", ab = "cipro")
plot(some_disk_values, mo = "Escherichia coli", ab = "cipro")
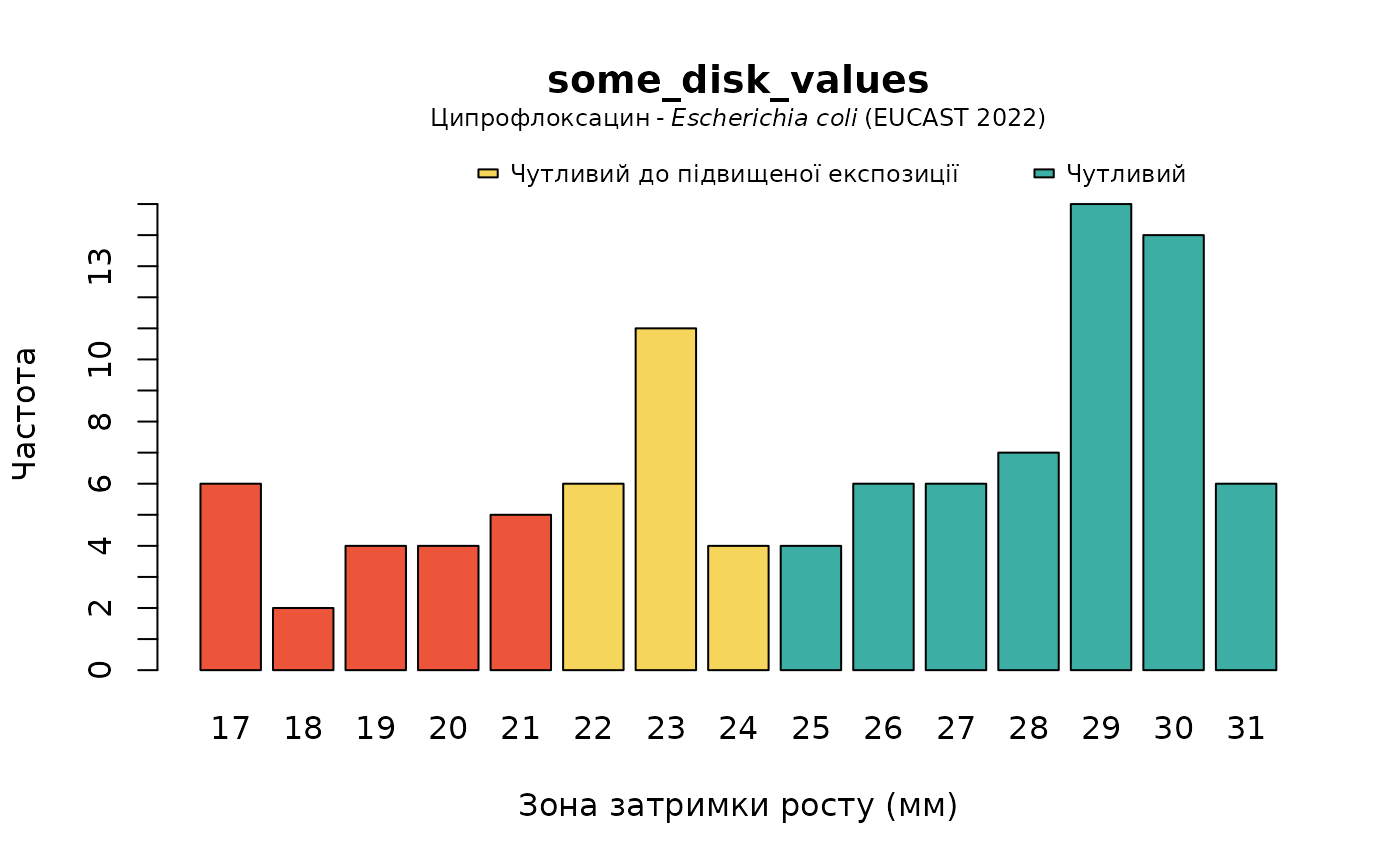
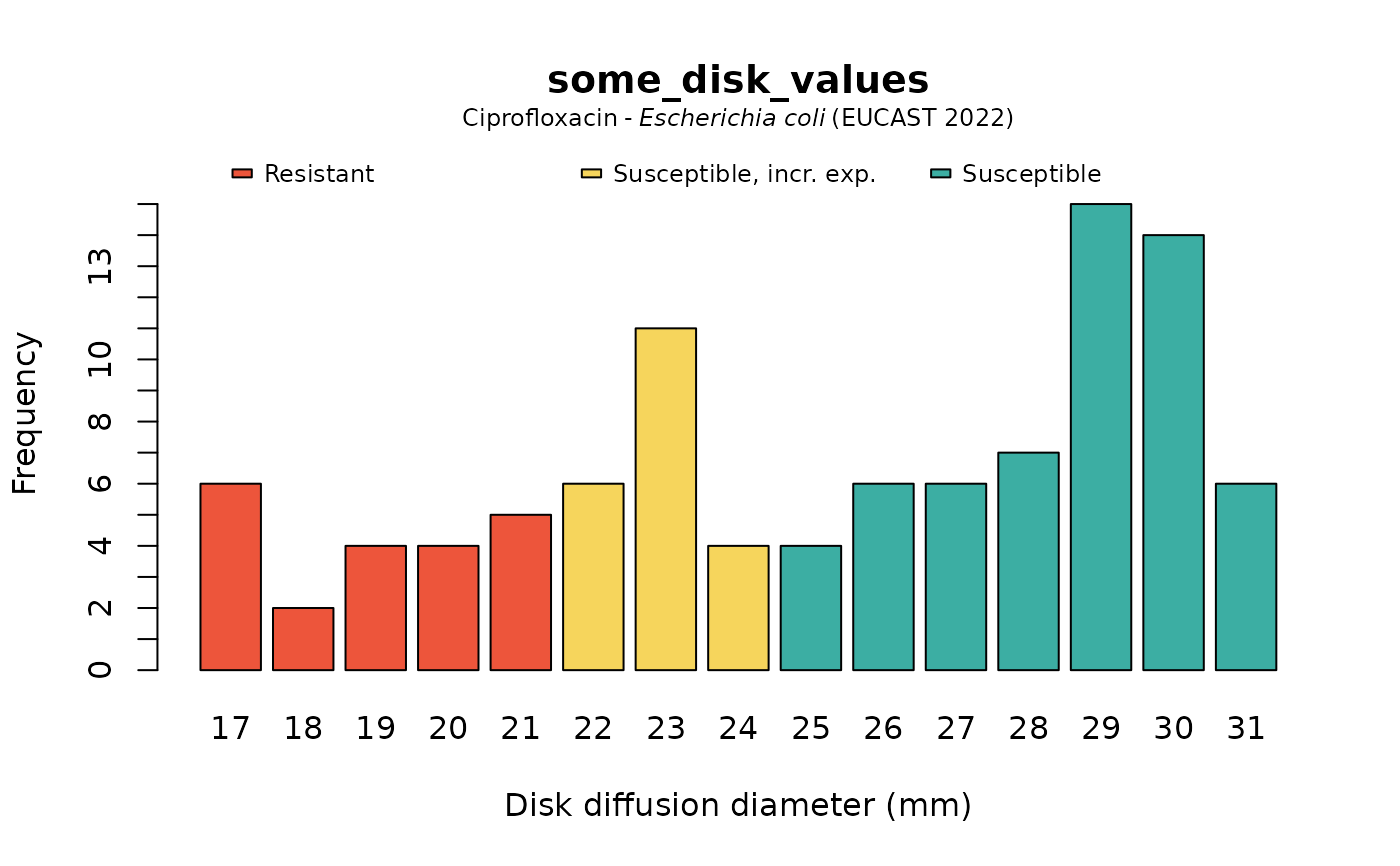 plot(some_disk_values, mo = "Escherichia coli", ab = "cipro", language = "uk")
plot(some_disk_values, mo = "Escherichia coli", ab = "cipro", language = "uk")
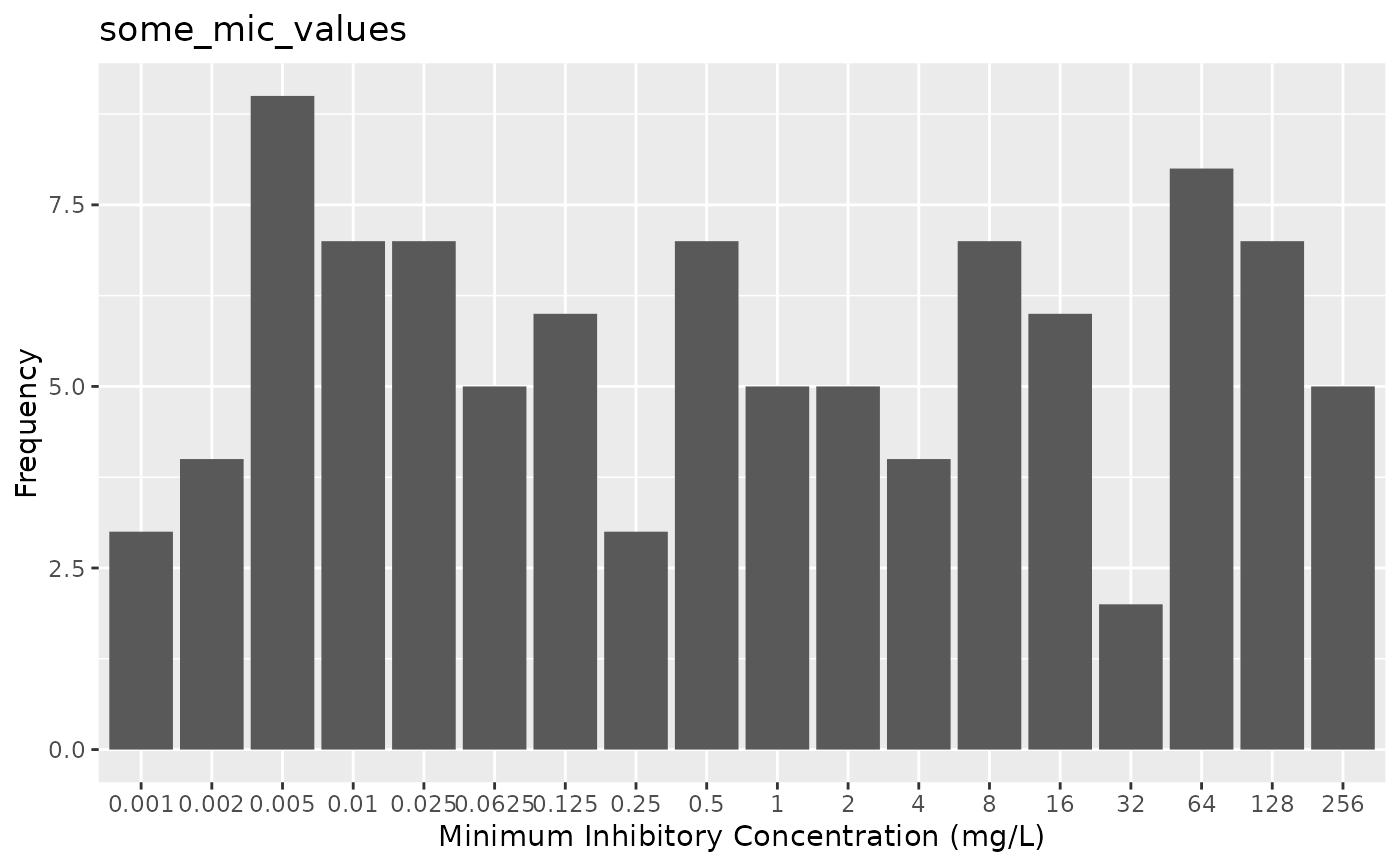
 # \donttest{
if (require("ggplot2")) {
autoplot(some_mic_values)
}
# \donttest{
if (require("ggplot2")) {
autoplot(some_mic_values)
}
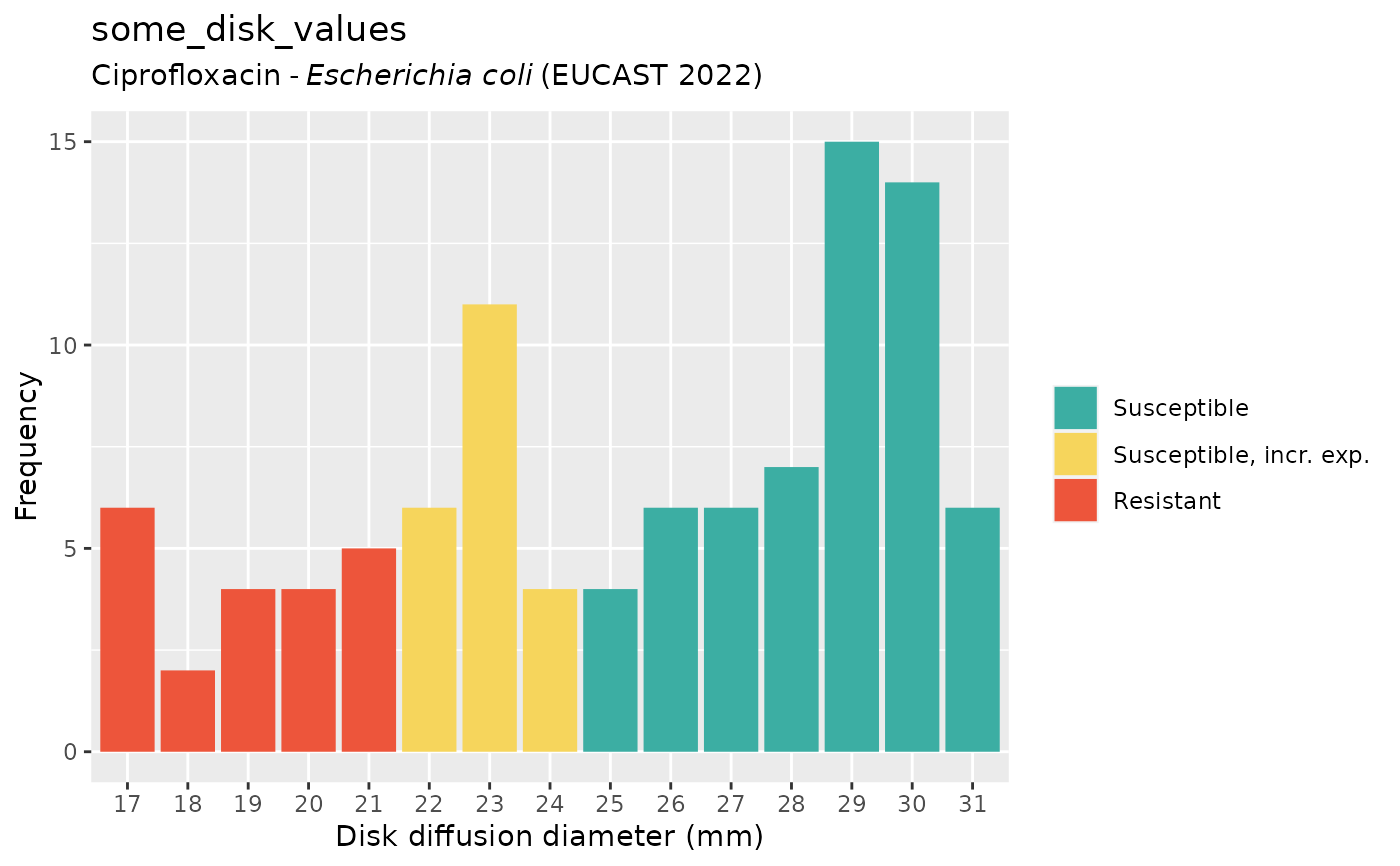
 if (require("ggplot2")) {
autoplot(some_disk_values, mo = "Escherichia coli", ab = "cipro")
}
if (require("ggplot2")) {
autoplot(some_disk_values, mo = "Escherichia coli", ab = "cipro")
}
 if (require("ggplot2")) {
autoplot(some_sir_values)
}
if (require("ggplot2")) {
autoplot(some_sir_values)
}
 # }
# }