One of the most important features of this package is the complete microbial taxonomic database, supplied by the Catalogue of Life (CoL) and the List of Prokaryotic names with Standing in Nomenclature (LPSN). We created a function as.mo() that transforms any user input value to a valid microbial ID by using intelligent rules combined with the microbial taxonomy.
Using the microbenchmark package, we can review the calculation performance of this function. Its function microbenchmark() runs different input expressions independently of each other and measures their time-to-result.
In the next test, we try to ‘coerce’ different input values into the microbial code of Staphylococcus aureus. Coercion is a computational process of forcing output based on an input. For microorganism names, coercing user input to taxonomically valid microorganism names is crucial to ensure correct interpretation and to enable grouping based on taxonomic properties.
The actual result is the same every time: it returns its microorganism code B_STPHY_AURS (B stands for Bacteria, its taxonomic kingdom).
But the calculation time differs a lot:
S.aureus <- microbenchmark(
as.mo("sau"), # WHONET code
as.mo("stau"),
as.mo("STAU"),
as.mo("staaur"),
as.mo("STAAUR"),
as.mo("S. aureus"),
as.mo("S aureus"),
as.mo("Staphylococcus aureus"), # official taxonomic name
as.mo("Staphylococcus aureus (MRSA)"), # additional text
as.mo("Sthafilokkockus aaureuz"), # incorrect spelling
as.mo("MRSA"), # Methicillin Resistant S. aureus
as.mo("VISA"), # Vancomycin Intermediate S. aureus
times = 25)
print(S.aureus, unit = "ms", signif = 2)
# Unit: milliseconds
# expr min lq mean median uq max neval
# as.mo("sau") 12.0 13.0 20.0 14.0 16.0 51 25
# as.mo("stau") 56.0 60.0 77.0 85.0 94.0 100 25
# as.mo("STAU") 54.0 59.0 78.0 85.0 95.0 100 25
# as.mo("staaur") 12.0 13.0 20.0 14.0 15.0 57 25
# as.mo("STAAUR") 13.0 13.0 19.0 14.0 16.0 51 25
# as.mo("S. aureus") 27.0 29.0 39.0 34.0 50.0 64 25
# as.mo("S aureus") 27.0 30.0 43.0 33.0 60.0 70 25
# as.mo("Staphylococcus aureus") 3.8 4.1 5.5 4.5 4.7 31 25
# as.mo("Staphylococcus aureus (MRSA)") 250.0 270.0 280.0 280.0 290.0 390 25
# as.mo("Sthafilokkockus aaureuz") 190.0 200.0 200.0 200.0 200.0 220 25
# as.mo("MRSA") 12.0 13.0 15.0 14.0 16.0 30 25
# as.mo("VISA") 21.0 22.0 31.0 25.0 29.0 59 25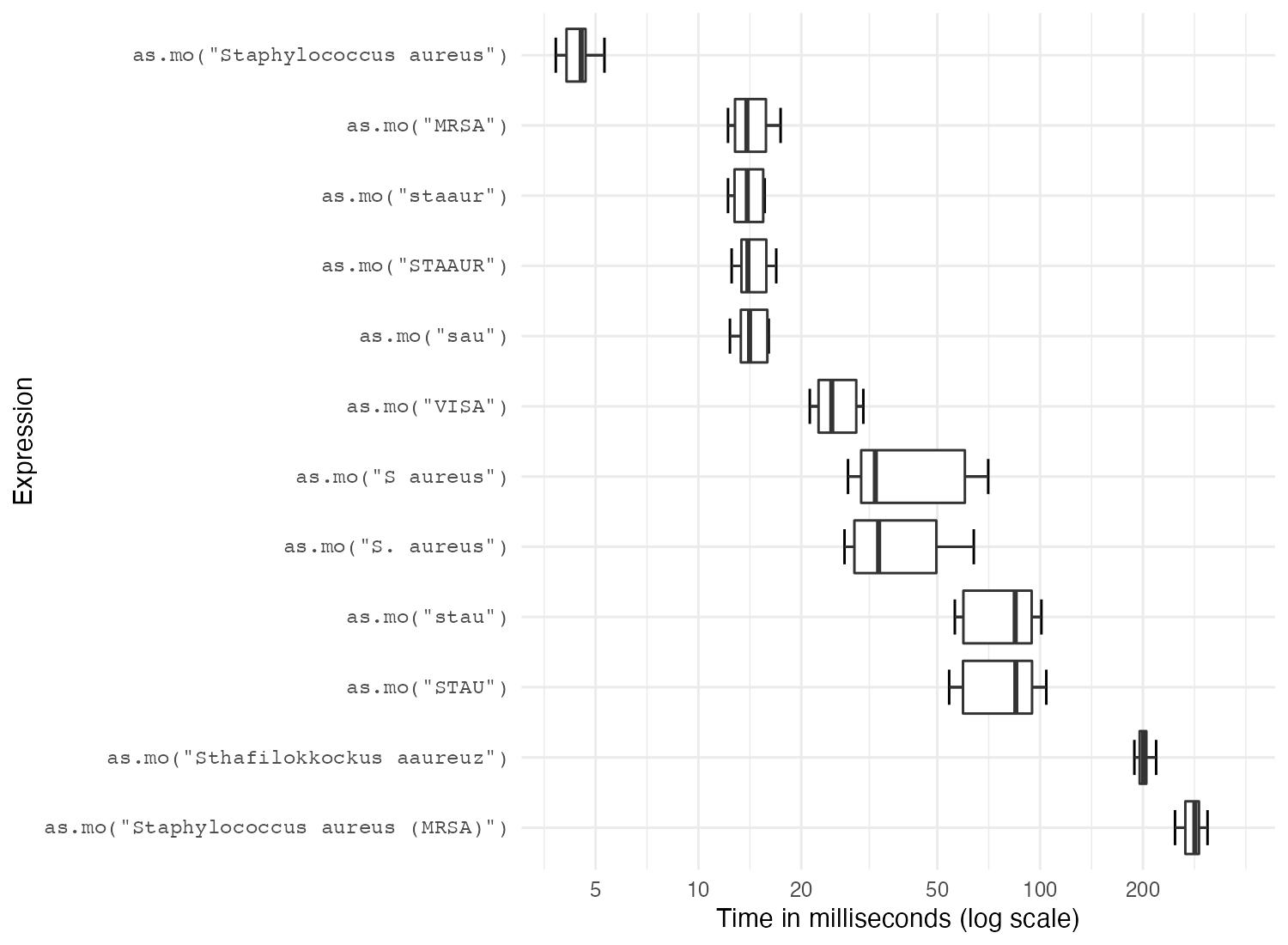
In the table above, all measurements are shown in milliseconds (thousands of seconds). A value of 5 milliseconds means it can determine 200 input values per second. It case of 200 milliseconds, this is only 5 input values per second. It is clear that accepted taxonomic names are extremely fast, but some variations are up to 200 times slower to determine.
To improve performance, we implemented two important algorithms to save unnecessary calculations: repetitive results and already precalculated results.
Repetitive results
Repetitive results are values that are present more than once in a vector. Unique values will only be calculated once by as.mo(). So running as.mo(c("E. coli", "E. coli")) will check the value "E. coli" only once.
To prove this, we will use mo_name() for testing - a helper function that returns the full microbial name (genus, species and possibly subspecies) which uses as.mo() internally.
# start with the example_isolates data set
x <- example_isolates %>%
# take all MO codes from the 'mo' column
pull(mo) %>%
# and copy them a thousand times
rep(1000) %>%
# then scramble them
sample()
# what do these values look like? They are of class <mo>:
head(x)
# Class <mo>
# [1] B_STPHY_CONS B_ESCHR_COLI B_STPHY_CONS B_ESCHR_COLI B_ESCHR_COLI
# [6] B_STPHY_HMNS
# as the example_isolates data set has 2,000 rows, we should have 2 million items
length(x)
# [1] 2000000
# and how many unique values do we have?
n_distinct(x)
# [1] 90
# now let's see:
run_it <- microbenchmark(mo_name(x),
times = 10)
print(run_it, unit = "ms", signif = 3)
# Unit: milliseconds
# expr min lq mean median uq max neval
# mo_name(x) 188 197 265 241 348 419 10So getting official taxonomic names of 2,000,000 (!!) items consisting of 90 unique values only takes 0.241 seconds. That is 121 nanoseconds on average. You only lose time on your unique input values.
Precalculated results
What about precalculated results? If the input is an already precalculated result of a helper function such as mo_name(), it almost doesn’t take any time at all. In other words, if you run mo_name() on a valid taxonomic name, it will return the results immediately (see ‘C’ below):
run_it <- microbenchmark(A = mo_name("STAAUR"),
B = mo_name("S. aureus"),
C = mo_name("Staphylococcus aureus"),
times = 10)
print(run_it, unit = "ms", signif = 3)
# Unit: milliseconds
# expr min lq mean median uq max neval
# A 7.98 8.26 15.80 8.35 9.35 60.00 10
# B 22.30 22.90 23.90 23.50 24.30 27.20 10
# C 1.74 2.02 2.16 2.16 2.24 2.65 10So going from mo_name("Staphylococcus aureus") to "Staphylococcus aureus" takes 0.0022 seconds - it doesn’t even start calculating if the result would be the same as the expected resulting value. That goes for all helper functions:
run_it <- microbenchmark(A = mo_species("aureus"),
B = mo_genus("Staphylococcus"),
C = mo_name("Staphylococcus aureus"),
D = mo_family("Staphylococcaceae"),
E = mo_order("Bacillales"),
F = mo_class("Bacilli"),
G = mo_phylum("Firmicutes"),
H = mo_kingdom("Bacteria"),
times = 10)
print(run_it, unit = "ms", signif = 3)
# Unit: milliseconds
# expr min lq mean median uq max neval
# A 1.65 1.67 1.74 1.69 1.83 1.89 10
# B 1.64 1.65 1.72 1.69 1.78 1.86 10
# C 1.67 1.75 1.84 1.80 1.87 2.19 10
# D 1.65 1.68 1.77 1.72 1.80 2.14 10
# E 1.65 1.68 1.97 1.75 2.18 2.89 10
# F 1.57 1.64 1.71 1.67 1.74 2.02 10
# G 1.62 1.67 1.79 1.68 1.72 2.71 10
# H 1.63 1.70 1.84 1.72 2.04 2.27 10Of course, when running mo_phylum("Firmicutes") the function has zero knowledge about the actual microorganism, namely S. aureus. But since the result would be "Firmicutes" anyway, there is no point in calculating the result. And because this package contains all phyla of all known bacteria, it can just return the initial value immediately.
Results in other languages
When the system language is non-English and supported by this AMR package, some functions will have a translated result. This almost does’t take extra time:
mo_name("CoNS", language = "en") # or just mo_name("CoNS") on an English system
# [1] "Coagulase-negative Staphylococcus (CoNS)"
mo_name("CoNS", language = "es") # or just mo_name("CoNS") on a Spanish system
# [1] "Staphylococcus coagulasa negativo (SCN)"
mo_name("CoNS", language = "nl") # or just mo_name("CoNS") on a Dutch system
# [1] "Coagulase-negatieve Staphylococcus (CNS)"
run_it <- microbenchmark(en = mo_name("CoNS", language = "en"),
de = mo_name("CoNS", language = "de"),
nl = mo_name("CoNS", language = "nl"),
es = mo_name("CoNS", language = "es"),
it = mo_name("CoNS", language = "it"),
fr = mo_name("CoNS", language = "fr"),
pt = mo_name("CoNS", language = "pt"),
times = 100)
print(run_it, unit = "ms", signif = 4)
# Unit: milliseconds
# expr min lq mean median uq max neval
# en 19.21 19.99 26.19 20.57 22.10 194.10 100
# de 30.22 31.42 42.42 32.36 35.01 94.70 100
# nl 34.40 35.36 43.21 36.35 38.65 119.80 100
# es 33.99 35.21 44.98 36.40 38.36 101.00 100
# it 23.55 24.66 27.67 25.19 26.54 76.79 100
# fr 23.47 24.32 27.80 25.02 26.51 82.05 100
# pt 23.43 24.36 30.42 24.94 26.09 80.48 100Currently supported non-English languages are German, Dutch, Spanish, Italian, French and Portuguese.