Interpret minimum inhibitory concentration (MIC) values and disk diffusion diameters according to EUCAST or CLSI, or clean up existing R/SI values. This transforms the input to a new class rsi, which is an ordered factor with levels S < I < R.
Usage
as.rsi(x, ...)
NA_rsi_
is.rsi(x)
is.rsi.eligible(x, threshold = 0.05)
# S3 method for mic
as.rsi(
x,
mo = NULL,
ab = deparse(substitute(x)),
guideline = "EUCAST",
uti = FALSE,
conserve_capped_values = FALSE,
add_intrinsic_resistance = FALSE,
reference_data = AMR::rsi_translation,
...
)
# S3 method for disk
as.rsi(
x,
mo = NULL,
ab = deparse(substitute(x)),
guideline = "EUCAST",
uti = FALSE,
add_intrinsic_resistance = FALSE,
reference_data = AMR::rsi_translation,
...
)
# S3 method for data.frame
as.rsi(
x,
...,
col_mo = NULL,
guideline = "EUCAST",
uti = NULL,
conserve_capped_values = FALSE,
add_intrinsic_resistance = FALSE,
reference_data = AMR::rsi_translation
)Arguments
- x
vector of values (for class
mic: MIC values in mg/L, for classdisk: a disk diffusion radius in millimetres)- ...
for using on a data.frame: names of columns to apply
as.rsi()on (supports tidy selection such ascolumn1:column4). Otherwise: arguments passed on to methods.- threshold
maximum fraction of invalid antimicrobial interpretations of
x, see Examples- mo
any (vector of) text that can be coerced to valid microorganism codes with
as.mo(), can be left empty to determine it automatically- ab
any (vector of) text that can be coerced to a valid antimicrobial code with
as.ab()- guideline
defaults to EUCAST 2022 (the latest implemented EUCAST guideline in the rsi_translation data set), supports EUCAST (2011-2022) and CLSI (2011-2022), see Details
- uti
(Urinary Tract Infection) A vector with logicals (
TRUEorFALSE) to specify whether a UTI specific interpretation from the guideline should be chosen. For usingas.rsi()on a data.frame, this can also be a column containing logicals or when left blank, the data set will be searched for a column 'specimen', and rows within this column containing 'urin' (such as 'urine', 'urina') will be regarded isolates from a UTI. See Examples.- conserve_capped_values
a logical to indicate that MIC values starting with
">"(but not">=") must always return "R" , and that MIC values starting with"<"(but not"<=") must always return "S"- add_intrinsic_resistance
(only useful when using a EUCAST guideline) a logical to indicate whether intrinsic antibiotic resistance must also be considered for applicable bug-drug combinations, meaning that e.g. ampicillin will always return "R" in Klebsiella species. Determination is based on the intrinsic_resistant data set, that itself is based on 'EUCAST Expert Rules' and 'EUCAST Intrinsic Resistance and Unusual Phenotypes' v3.3 (2021).
- reference_data
a data.frame to be used for interpretation, which defaults to the rsi_translation data set. Changing this argument allows for using own interpretation guidelines. This argument must contain a data set that is equal in structure to the rsi_translation data set (same column names and column types). Please note that the
guidelineargument will be ignored whenreference_datais manually set.- col_mo
column name of the IDs of the microorganisms (see
as.mo()), defaults to the first column of classmo. Values will be coerced usingas.mo().
Value
Ordered factor with new class <rsi>
Details
How it Works
The as.rsi() function works in four ways:
For cleaning raw / untransformed data. The data will be cleaned to only contain values S, I and R and will try its best to determine this with some intelligence. For example, mixed values with R/SI interpretations and MIC values such as
"<0.25; S"will be coerced to"S". Combined interpretations for multiple test methods (as seen in laboratory records) such as"S; S"will be coerced to"S", but a value like"S; I"will returnNAwith a warning that the input is unclear.For interpreting minimum inhibitory concentration (MIC) values according to EUCAST or CLSI. You must clean your MIC values first using
as.mic(), that also gives your columns the new data classmic. Also, be sure to have a column with microorganism names or codes. It will be found automatically, but can be set manually using themoargument.Using
dplyr, R/SI interpretation can be done very easily with either:your_data %>% mutate_if(is.mic, as.rsi) # until dplyr 1.0.0 your_data %>% mutate(across(where(is.mic), as.rsi)) # since dplyr 1.0.0Operators like "<=" will be stripped before interpretation. When using
conserve_capped_values = TRUE, an MIC value of e.g. ">2" will always return "R", even if the breakpoint according to the chosen guideline is ">=4". This is to prevent that capped values from raw laboratory data would not be treated conservatively. The default behaviour (conserve_capped_values = FALSE) considers ">2" to be lower than ">=4" and might in this case return "S" or "I".
For interpreting disk diffusion diameters according to EUCAST or CLSI. You must clean your disk zones first using
as.disk(), that also gives your columns the new data classdisk. Also, be sure to have a column with microorganism names or codes. It will be found automatically, but can be set manually using themoargument.Using
dplyr, R/SI interpretation can be done very easily with either:your_data %>% mutate_if(is.disk, as.rsi) # until dplyr 1.0.0 your_data %>% mutate(across(where(is.disk), as.rsi)) # since dplyr 1.0.0
For interpreting a complete data set, with automatic determination of MIC values, disk diffusion diameters, microorganism names or codes, and antimicrobial test results. This is done very simply by running
as.rsi(your_data).
Supported Guidelines
For interpreting MIC values as well as disk diffusion diameters, currently implemented guidelines are EUCAST (2011-2022) and CLSI (2011-2022).
Thus, the guideline argument must be set to e.g., "EUCAST 2022" or "CLSI 2022". By simply using "EUCAST" (the default) or "CLSI" as input, the latest included version of that guideline will automatically be selected. You can set your own data set using the reference_data argument. The guideline argument will then be ignored.
After Interpretation
After using as.rsi(), you can use the eucast_rules() defined by EUCAST to (1) apply inferred susceptibility and resistance based on results of other antimicrobials and (2) apply intrinsic resistance based on taxonomic properties of a microorganism.
Machine-Readable Interpretation Guidelines
The repository of this package contains a machine-readable version of all guidelines. This is a CSV file consisting of 20,369 rows and 11 columns. This file is machine-readable, since it contains one row for every unique combination of the test method (MIC or disk diffusion), the antimicrobial agent and the microorganism. This allows for easy implementation of these rules in laboratory information systems (LIS). Note that it only contains interpretation guidelines for humans - interpretation guidelines from CLSI for animals were removed.
Other
The function is.rsi() detects if the input contains class <rsi>. If the input is a data.frame, it iterates over all columns and returns a logical vector.
The function is.rsi.eligible() returns TRUE when a columns contains at most 5% invalid antimicrobial interpretations (not S and/or I and/or R), and FALSE otherwise. The threshold of 5% can be set with the threshold argument. If the input is a data.frame, it iterates over all columns and returns a logical vector.
NA_rsi_ is a missing value of the new <rsi> class, analogous to e.g. base R's NA_character_.
Interpretation of R and S/I
In 2019, the European Committee on Antimicrobial Susceptibility Testing (EUCAST) has decided to change the definitions of susceptibility testing categories R and S/I as shown below (https://www.eucast.org/newsiandr/).
R = Resistant
A microorganism is categorised as Resistant when there is a high likelihood of therapeutic failure even when there is increased exposure. Exposure is a function of how the mode of administration, dose, dosing interval, infusion time, as well as distribution and excretion of the antimicrobial agent will influence the infecting organism at the site of infection.S = Susceptible
A microorganism is categorised as Susceptible, standard dosing regimen, when there is a high likelihood of therapeutic success using a standard dosing regimen of the agent.I = Susceptible, Increased exposure
A microorganism is categorised as Susceptible, Increased exposure when there is a high likelihood of therapeutic success because exposure to the agent is increased by adjusting the dosing regimen or by its concentration at the site of infection.
This AMR package honours this (new) insight. Use susceptibility() (equal to proportion_SI()) to determine antimicrobial susceptibility and count_susceptible() (equal to count_SI()) to count susceptible isolates.
Reference Data Publicly Available
All reference data sets (about microorganisms, antibiotics, R/SI interpretation, EUCAST rules, etc.) in this AMR package are publicly and freely available. We continually export our data sets to formats for use in R, SPSS, SAS, Stata and Excel. We also supply flat files that are machine-readable and suitable for input in any software program, such as laboratory information systems. Please find all download links on our website, which is automatically updated with every code change.
Examples
example_isolates
#> # A tibble: 2,000 × 49
#> date hospit…¹ ward_…² ward_…³ ward_…⁴ age gender patie…⁵ mo
#> <date> <fct> <lgl> <lgl> <lgl> <dbl> <chr> <chr> <mo>
#> 1 2002-01-02 D FALSE TRUE FALSE 65 F A77334 B_ESCHR_COLI
#> 2 2002-01-03 D FALSE TRUE FALSE 65 F A77334 B_ESCHR_COLI
#> 3 2002-01-07 B TRUE FALSE FALSE 45 F 067927 B_STPHY_EPDR
#> 4 2002-01-07 B TRUE FALSE FALSE 45 F 067927 B_STPHY_EPDR
#> 5 2002-01-13 B TRUE FALSE FALSE 45 F 067927 B_STPHY_EPDR
#> 6 2002-01-13 B TRUE FALSE FALSE 45 F 067927 B_STPHY_EPDR
#> 7 2002-01-14 D FALSE TRUE FALSE 78 M 462729 B_STPHY_AURS
#> 8 2002-01-14 D FALSE TRUE FALSE 78 M 462729 B_STPHY_AURS
#> 9 2002-01-16 B TRUE FALSE FALSE 45 F 067927 B_STPHY_EPDR
#> 10 2002-01-17 B TRUE FALSE FALSE 79 F 858515 B_STPHY_EPDR
#> # … with 1,990 more rows, 40 more variables: PEN <rsi>, OXA <rsi>, FLC <rsi>,
#> # AMX <rsi>, AMC <rsi>, AMP <rsi>, TZP <rsi>, CZO <rsi>, FEP <rsi>,
#> # CXM <rsi>, FOX <rsi>, CTX <rsi>, CAZ <rsi>, CRO <rsi>, GEN <rsi>,
#> # TOB <rsi>, AMK <rsi>, KAN <rsi>, TMP <rsi>, SXT <rsi>, NIT <rsi>,
#> # FOS <rsi>, LNZ <rsi>, CIP <rsi>, MFX <rsi>, VAN <rsi>, TEC <rsi>,
#> # TCY <rsi>, TGC <rsi>, DOX <rsi>, ERY <rsi>, CLI <rsi>, AZM <rsi>,
#> # IPM <rsi>, MEM <rsi>, MTR <rsi>, CHL <rsi>, COL <rsi>, MUP <rsi>, …
#> # ℹ Use `print(n = ...)` to see more rows, and `colnames()` to see all variable names
summary(example_isolates) # see all R/SI results at a glance
#> date hospital_id ward_icu ward_clinical
#> Min. :2002-01-02 A:321 Mode :logical Mode :logical
#> 1st Qu.:2005-07-31 B:663 FALSE:1354 FALSE:536
#> Median :2009-07-31 C:254 TRUE :646 TRUE :1464
#> Mean :2009-11-20 D:762
#> 3rd Qu.:2014-05-30
#> Max. :2017-12-28
#> ward_outpatient age gender patient_id
#> Mode :logical Min. : 0.00 Length:2000 Length:2000
#> FALSE:1880 1st Qu.:63.00 Class :character Class :character
#> TRUE :120 Median :74.00 Mode :character Mode :character
#> Mean :70.69
#> 3rd Qu.:82.00
#> Max. :97.00
#> mo PEN OXA
#> Class :mo Class:rsi Class:rsi
#> <NA> :0 %R :73.7% (n=1201) %R :31.2% (n=114)
#> Unique:90 %SI :26.3% (n=428) %SI :68.8% (n=251)
#> #1 :B_ESCHR_COLI - %S :25.6% (n=417) - %S :68.8% (n=251)
#> #2 :B_STPHY_CONS - %I : 0.7% (n=11) - %I : 0.0% (n=0)
#> #3 :B_STPHY_AURS
#> FLC AMX AMC
#> Class:rsi Class:rsi Class:rsi
#> %R :29.5% (n=278) %R :59.6% (n=804) %R :23.7% (n=446)
#> %SI :70.5% (n=665) %SI :40.4% (n=546) %SI :76.3% (n=1433)
#> - %S :70.5% (n=665) - %S :40.2% (n=543) - %S :71.4% (n=1342)
#> - %I : 0.0% (n=0) - %I : 0.2% (n=3) - %I : 4.8% (n=91)
#>
#> AMP TZP CZO
#> Class:rsi Class:rsi Class:rsi
#> %R :59.6% (n=804) %R :12.6% (n=126) %R :44.6% (n=199)
#> %SI :40.4% (n=546) %SI :87.4% (n=875) %SI :55.4% (n=247)
#> - %S :40.2% (n=543) - %S :86.1% (n=862) - %S :54.9% (n=245)
#> - %I : 0.2% (n=3) - %I : 1.3% (n=13) - %I : 0.4% (n=2)
#>
#> FEP CXM FOX
#> Class:rsi Class:rsi Class:rsi
#> %R :14.2% (n=103) %R :26.3% (n=470) %R :27.4% (n=224)
#> %SI :85.8% (n=621) %SI :73.7% (n=1319) %SI :72.6% (n=594)
#> - %S :85.6% (n=620) - %S :72.5% (n=1297) - %S :71.6% (n=586)
#> - %I : 0.1% (n=1) - %I : 1.2% (n=22) - %I : 1.0% (n=8)
#>
#> CTX CAZ CRO
#> Class:rsi Class:rsi Class:rsi
#> %R :15.5% (n=146) %R :66.5% (n=1204) %R :15.5% (n=146)
#> %SI :84.5% (n=797) %SI :33.5% (n=607) %SI :84.5% (n=797)
#> - %S :84.4% (n=796) - %S :33.5% (n=607) - %S :84.4% (n=796)
#> - %I : 0.1% (n=1) - %I : 0.0% (n=0) - %I : 0.1% (n=1)
#>
#> GEN TOB AMK
#> Class:rsi Class:rsi Class:rsi
#> %R :24.6% (n=456) %R :34.4% (n=465) %R :63.7% (n=441)
#> %SI :75.4% (n=1399) %SI :65.6% (n=886) %SI :36.3% (n=251)
#> - %S :74.0% (n=1372) - %S :65.1% (n=879) - %S :36.3% (n=251)
#> - %I : 1.5% (n=27) - %I : 0.5% (n=7) - %I : 0.0% (n=0)
#>
#> KAN TMP SXT
#> Class:rsi Class:rsi Class:rsi
#> %R :100.0% (n=471) %R :38.1% (n=571) %R :20.5% (n=361)
#> %SI : 0.0% (n=0) %SI :61.9% (n=928) %SI :79.5% (n=1398)
#> - %S : 0.0% (n=0) - %S :61.2% (n=918) - %S :79.1% (n=1392)
#> - %I : 0.0% (n=0) - %I : 0.7% (n=10) - %I : 0.3% (n=6)
#>
#> NIT FOS LNZ
#> Class:rsi Class:rsi Class:rsi
#> %R :17.1% (n=127) %R :42.2% (n=148) %R :69.3% (n=709)
#> %SI :82.9% (n=616) %SI :57.8% (n=203) %SI :30.7% (n=314)
#> - %S :76.0% (n=565) - %S :57.8% (n=203) - %S :30.7% (n=314)
#> - %I : 6.9% (n=51) - %I : 0.0% (n=0) - %I : 0.0% (n=0)
#>
#> CIP MFX VAN
#> Class:rsi Class:rsi Class:rsi
#> %R :16.2% (n=228) %R :33.6% (n=71) %R :38.3% (n=712)
#> %SI :83.8% (n=1181) %SI :66.4% (n=140) %SI :61.7% (n=1149)
#> - %S :78.9% (n=1112) - %S :64.5% (n=136) - %S :61.7% (n=1149)
#> - %I : 4.9% (n=69) - %I : 1.9% (n=4) - %I : 0.0% (n=0)
#>
#> TEC TCY TGC
#> Class:rsi Class:rsi Class:rsi
#> %R :75.7% (n=739) %R :29.8% (n=357) %R :12.7% (n=101)
#> %SI :24.3% (n=237) %SI :70.3% (n=843) %SI :87.3% (n=697)
#> - %S :24.3% (n=237) - %S :68.3% (n=820) - %S :87.3% (n=697)
#> - %I : 0.0% (n=0) - %I : 1.9% (n=23) - %I : 0.0% (n=0)
#>
#> DOX ERY CLI
#> Class:rsi Class:rsi Class:rsi
#> %R :27.7% (n=315) %R :57.2% (n=1084) %R :61.2% (n=930)
#> %SI :72.3% (n=821) %SI :42.8% (n=810) %SI :38.8% (n=590)
#> - %S :71.7% (n=814) - %S :42.3% (n=801) - %S :38.6% (n=586)
#> - %I : 0.6% (n=7) - %I : 0.5% (n=9) - %I : 0.3% (n=4)
#>
#> AZM IPM MEM
#> Class:rsi Class:rsi Class:rsi
#> %R :57.2% (n=1084) %R : 6.2% (n=55) %R : 5.9% (n=49)
#> %SI :42.8% (n=810) %SI :93.8% (n=834) %SI :94.1% (n=780)
#> - %S :42.3% (n=801) - %S :92.7% (n=824) - %S :94.1% (n=780)
#> - %I : 0.5% (n=9) - %I : 1.1% (n=10) - %I : 0.0% (n=0)
#>
#> MTR CHL COL
#> Class:rsi Class:rsi Class:rsi
#> %R :14.7% (n=5) %R :21.4% (n=33) %R :81.2% (n=1331)
#> %SI :85.3% (n=29) %SI :78.6% (n=121) %SI :18.8% (n=309)
#> - %S :85.3% (n=29) - %S :78.6% (n=121) - %S :18.8% (n=309)
#> - %I : 0.0% (n=0) - %I : 0.0% (n=0) - %I : 0.0% (n=0)
#>
#> MUP RIF
#> Class:rsi Class:rsi
#> %R : 5.9% (n=16) %R :69.6% (n=698)
#> %SI :94.1% (n=254) %SI :30.4% (n=305)
#> - %S :93.0% (n=251) - %S :30.2% (n=303)
#> - %I : 1.1% (n=3) - %I : 0.2% (n=2)
#>
# For INTERPRETING disk diffusion and MIC values -----------------------
# a whole data set, even with combined MIC values and disk zones
df <- data.frame(microorganism = "Escherichia coli",
AMP = as.mic(8),
CIP = as.mic(0.256),
GEN = as.disk(18),
TOB = as.disk(16),
NIT = as.mic(32),
ERY = "R")
as.rsi(df)
#> ℹ Using column 'microorganism' as input for `col_mo`.
#> => Interpreting MIC values of column 'AMP' (AMP, ampicillin) according to
#> EUCAST 2022...
#> OK.
#> => Interpreting MIC values of column 'CIP' (CIP, ciprofloxacin) according
#> to EUCAST 2022...
#> OK.
#> => Interpreting disk diffusion zones of column 'GEN' (GEN, gentamicin)
#> according to EUCAST 2022...
#> Warning: in `as.rsi()`: interpretation of gentamicin is only available for
#> (uncomplicated) urinary tract infections (UTI) for some microorganisms. Use
#> argument `uti` to set which isolates are from urine. See ?as.rsi.
#> WARNING.
#> => Interpreting disk diffusion zones of column 'TOB' (TOB, tobramycin)
#> according to EUCAST 2022...
#> Warning: in `as.rsi()`: interpretation of tobramycin is only available for
#> (uncomplicated) urinary tract infections (UTI) for some microorganisms. Use
#> argument `uti` to set which isolates are from urine. See ?as.rsi.
#> WARNING.
#> => Interpreting MIC values of column 'NIT' (NIT, nitrofurantoin) according
#> to EUCAST 2022...
#> Warning: in `as.rsi()`: interpretation of nitrofurantoin is only available for
#> (uncomplicated) urinary tract infections (UTI) for some microorganisms. Use
#> argument `uti` to set which isolates are from urine. See ?as.rsi.
#> WARNING.
#> => Assigning class <rsi> to already clean column 'ERY' (erythromycin)...
#> OK.
#> microorganism AMP CIP GEN TOB NIT ERY
#> 1 Escherichia coli S I S S <NA> R
# for single values
as.rsi(x = as.mic(2),
mo = as.mo("S. pneumoniae"),
ab = "AMP",
guideline = "EUCAST")
#> ℹ Function `as.mo()` is uncertain about "S. pneumoniae" (assuming
#> Streptococcus pneumoniae). Run `mo_uncertainties()` to review this.
#> => Interpreting MIC values of 'AMP' (AMP, ampicillin) according to EUCAST
#> 2022...
#> OK.
#> Class <rsi>
#> [1] R
as.rsi(x = as.disk(18),
mo = "Strep pneu", # `mo` will be coerced with as.mo()
ab = "ampicillin", # and `ab` with as.ab()
guideline = "EUCAST")
#> => Interpreting disk diffusion zones of 'ampicillin' according to EUCAST
#> 2022...
#> OK.
#> Class <rsi>
#> [1] R
# \donttest{
# the dplyr way
if (require("dplyr")) {
df %>% mutate_if(is.mic, as.rsi)
df %>% mutate_if(function(x) is.mic(x) | is.disk(x), as.rsi)
df %>% mutate(across(where(is.mic), as.rsi))
df %>% mutate_at(vars(AMP:TOB), as.rsi)
df %>% mutate(across(AMP:TOB, as.rsi))
df %>%
mutate_at(vars(AMP:TOB), as.rsi, mo = .$microorganism)
# to include information about urinary tract infections (UTI)
data.frame(mo = "E. coli",
NIT = c("<= 2", 32),
from_the_bladder = c(TRUE, FALSE)) %>%
as.rsi(uti = "from_the_bladder")
data.frame(mo = "E. coli",
NIT = c("<= 2", 32),
specimen = c("urine", "blood")) %>%
as.rsi() # automatically determines urine isolates
df %>%
mutate_at(vars(AMP:NIT), as.rsi, mo = "E. coli", uti = TRUE)
}
#> => Interpreting MIC values of 'AMP' (AMP, ampicillin) based on column
#> 'microorganism' according to EUCAST 2022...
#> OK.
#> => Interpreting MIC values of 'CIP' (CIP, ciprofloxacin) based on column
#> 'microorganism' according to EUCAST 2022...
#> OK.
#> => Interpreting MIC values of 'NIT' (NIT, nitrofurantoin) based on column
#> 'microorganism' according to EUCAST 2022...
#> Warning: in `as.rsi()`: interpretation of nitrofurantoin is only available for
#> (uncomplicated) urinary tract infections (UTI) for some microorganisms. Use
#> argument `uti` to set which isolates are from urine. See ?as.rsi.
#> WARNING.
#> => Interpreting MIC values of 'AMP' (AMP, ampicillin) based on column
#> 'microorganism' according to EUCAST 2022...
#> OK.
#> => Interpreting MIC values of 'CIP' (CIP, ciprofloxacin) based on column
#> 'microorganism' according to EUCAST 2022...
#> OK.
#> => Interpreting disk diffusion zones of 'GEN' (GEN, gentamicin) based on
#> column 'microorganism' according to EUCAST 2022...
#> Warning: in `as.rsi()`: interpretation of gentamicin is only available for
#> (uncomplicated) urinary tract infections (UTI) for some microorganisms. Use
#> argument `uti` to set which isolates are from urine. See ?as.rsi.
#> WARNING.
#> => Interpreting disk diffusion zones of 'TOB' (TOB, tobramycin) based on
#> column 'microorganism' according to EUCAST 2022...
#> Warning: in `as.rsi()`: interpretation of tobramycin is only available for
#> (uncomplicated) urinary tract infections (UTI) for some microorganisms. Use
#> argument `uti` to set which isolates are from urine. See ?as.rsi.
#> WARNING.
#> => Interpreting MIC values of 'NIT' (NIT, nitrofurantoin) based on column
#> 'microorganism' according to EUCAST 2022...
#> Warning: in `as.rsi()`: interpretation of nitrofurantoin is only available for
#> (uncomplicated) urinary tract infections (UTI) for some microorganisms. Use
#> argument `uti` to set which isolates are from urine. See ?as.rsi.
#> WARNING.
#> => Interpreting MIC values of 'AMP' (AMP, ampicillin) based on column
#> 'microorganism' according to EUCAST 2022...
#> OK.
#> => Interpreting MIC values of 'CIP' (CIP, ciprofloxacin) based on column
#> 'microorganism' according to EUCAST 2022...
#> OK.
#> => Interpreting MIC values of 'NIT' (NIT, nitrofurantoin) based on column
#> 'microorganism' according to EUCAST 2022...
#> Warning: in `as.rsi()`: interpretation of nitrofurantoin is only available for
#> (uncomplicated) urinary tract infections (UTI) for some microorganisms. Use
#> argument `uti` to set which isolates are from urine. See ?as.rsi.
#> WARNING.
#> => Interpreting MIC values of 'AMP' (AMP, ampicillin) based on column
#> 'microorganism' according to EUCAST 2022...
#> OK.
#> => Interpreting MIC values of 'CIP' (CIP, ciprofloxacin) based on column
#> 'microorganism' according to EUCAST 2022...
#> OK.
#> => Interpreting disk diffusion zones of 'GEN' (GEN, gentamicin) based on
#> column 'microorganism' according to EUCAST 2022...
#> Warning: in `as.rsi()`: interpretation of gentamicin is only available for
#> (uncomplicated) urinary tract infections (UTI) for some microorganisms. Use
#> argument `uti` to set which isolates are from urine. See ?as.rsi.
#> WARNING.
#> => Interpreting disk diffusion zones of 'TOB' (TOB, tobramycin) based on
#> column 'microorganism' according to EUCAST 2022...
#> Warning: in `as.rsi()`: interpretation of tobramycin is only available for
#> (uncomplicated) urinary tract infections (UTI) for some microorganisms. Use
#> argument `uti` to set which isolates are from urine. See ?as.rsi.
#> WARNING.
#> => Interpreting MIC values of 'AMP' (AMP, ampicillin) based on column
#> 'microorganism' according to EUCAST 2022...
#> OK.
#> => Interpreting MIC values of 'CIP' (CIP, ciprofloxacin) based on column
#> 'microorganism' according to EUCAST 2022...
#> OK.
#> => Interpreting disk diffusion zones of 'GEN' (GEN, gentamicin) based on
#> column 'microorganism' according to EUCAST 2022...
#> Warning: in `as.rsi()`: interpretation of gentamicin is only available for
#> (uncomplicated) urinary tract infections (UTI) for some microorganisms. Use
#> argument `uti` to set which isolates are from urine. See ?as.rsi.
#> WARNING.
#> => Interpreting disk diffusion zones of 'TOB' (TOB, tobramycin) based on
#> column 'microorganism' according to EUCAST 2022...
#> Warning: in `as.rsi()`: interpretation of tobramycin is only available for
#> (uncomplicated) urinary tract infections (UTI) for some microorganisms. Use
#> argument `uti` to set which isolates are from urine. See ?as.rsi.
#> WARNING.
#> => Interpreting MIC values of 'AMP' (AMP, ampicillin) according to EUCAST
#> 2022...
#> OK.
#> => Interpreting MIC values of 'CIP' (CIP, ciprofloxacin) according to
#> EUCAST 2022...
#> OK.
#> => Interpreting disk diffusion zones of 'GEN' (GEN, gentamicin) according
#> to EUCAST 2022...
#> Warning: in `as.rsi()`: interpretation of gentamicin is only available for
#> (uncomplicated) urinary tract infections (UTI) for some microorganisms. Use
#> argument `uti` to set which isolates are from urine. See ?as.rsi.
#> WARNING.
#> => Interpreting disk diffusion zones of 'TOB' (TOB, tobramycin) according
#> to EUCAST 2022...
#> Warning: in `as.rsi()`: interpretation of tobramycin is only available for
#> (uncomplicated) urinary tract infections (UTI) for some microorganisms. Use
#> argument `uti` to set which isolates are from urine. See ?as.rsi.
#> WARNING.
#> ℹ Using column 'mo' as input for `col_mo`.
#> ℹ Function `as.mo()` is uncertain about "E. coli" (assuming Escherichia
#> coli). Run `mo_uncertainties()` to review this.
#> => Interpreting MIC values of column 'NIT' (NIT, nitrofurantoin) according
#> to EUCAST 2022...
#> OK.
#> ℹ Assuming value "urine" in column 'specimen' reflects a urinary tract
#> infection.
#> Use `as.rsi(uti = FALSE)` to prevent this.
#> ℹ Using column 'mo' as input for `col_mo`.
#> ℹ Function `as.mo()` is uncertain about "E. coli" (assuming Escherichia
#> coli). Run `mo_uncertainties()` to review this.
#> => Interpreting MIC values of column 'NIT' (NIT, nitrofurantoin) according
#> to EUCAST 2022...
#> OK.
#> ℹ Function `as.mo()` is uncertain about "E. coli" (assuming Escherichia
#> coli). Run `mo_uncertainties()` to review this.
#> => Interpreting MIC values of 'AMP' (AMP, ampicillin) according to EUCAST
#> 2022...
#> OK.
#> => Interpreting MIC values of 'CIP' (CIP, ciprofloxacin) according to
#> EUCAST 2022...
#> OK.
#> => Interpreting disk diffusion zones of 'GEN' (GEN, gentamicin) according
#> to EUCAST 2022...
#> OK.
#> => Interpreting disk diffusion zones of 'TOB' (TOB, tobramycin) according
#> to EUCAST 2022...
#> OK.
#> => Interpreting MIC values of 'NIT' (NIT, nitrofurantoin) according to
#> EUCAST 2022...
#> OK.
#> microorganism AMP CIP GEN TOB NIT ERY
#> 1 Escherichia coli S I S S S R
# For CLEANING existing R/SI values ------------------------------------
as.rsi(c("S", "I", "R", "A", "B", "C"))
#> Warning: in `as.rsi()`: 3 results truncated (50%) that were invalid antimicrobial
#> interpretations: "A", "B" and "C"
#> Class <rsi>
#> [1] S I R <NA> <NA> <NA>
as.rsi("<= 0.002; S") # will return "S"
#> Class <rsi>
#> [1] S
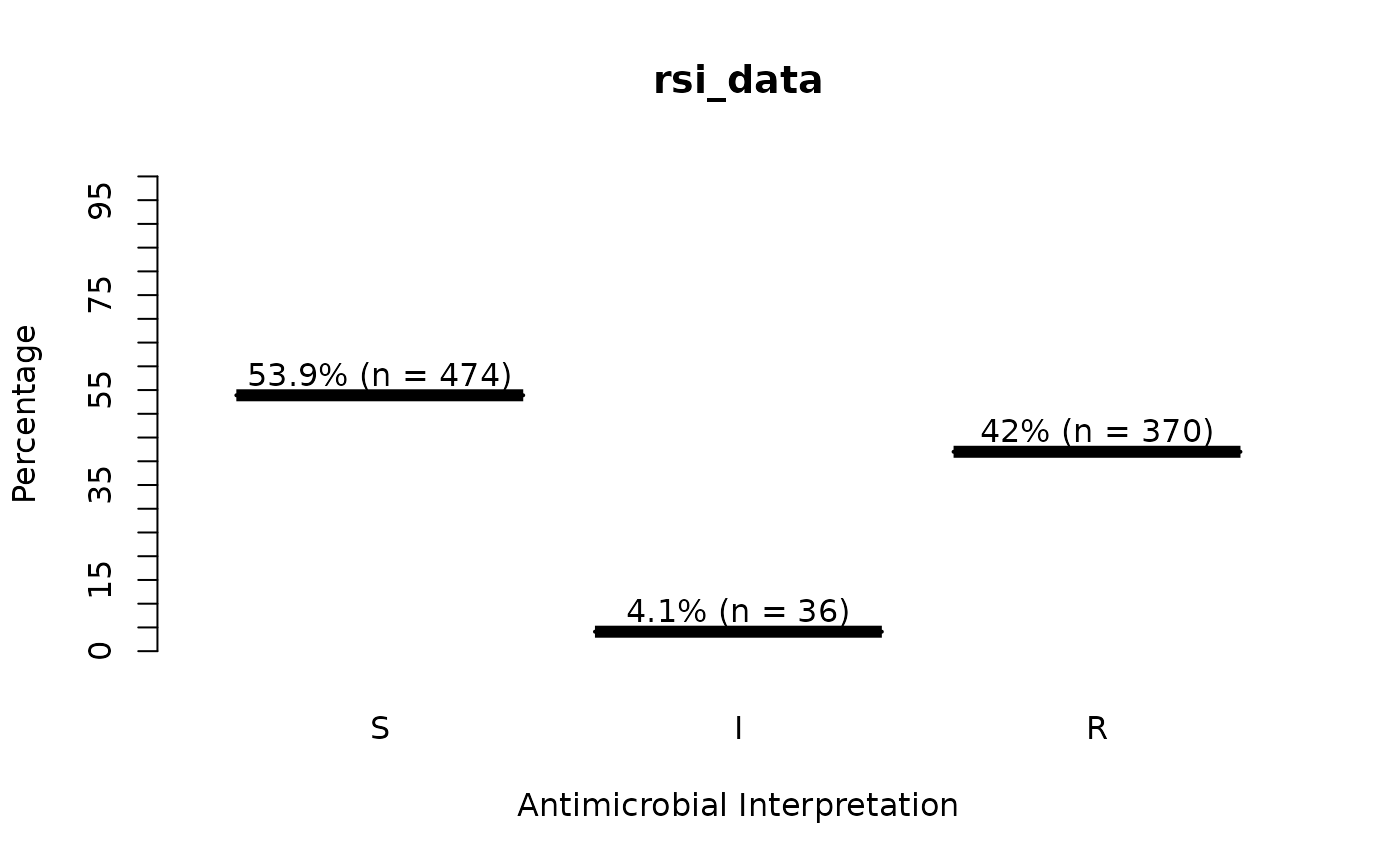
rsi_data <- as.rsi(c(rep("S", 474), rep("I", 36), rep("R", 370)))
is.rsi(rsi_data)
#> [1] TRUE
plot(rsi_data) # for percentages
 barplot(rsi_data) # for frequencies
barplot(rsi_data) # for frequencies
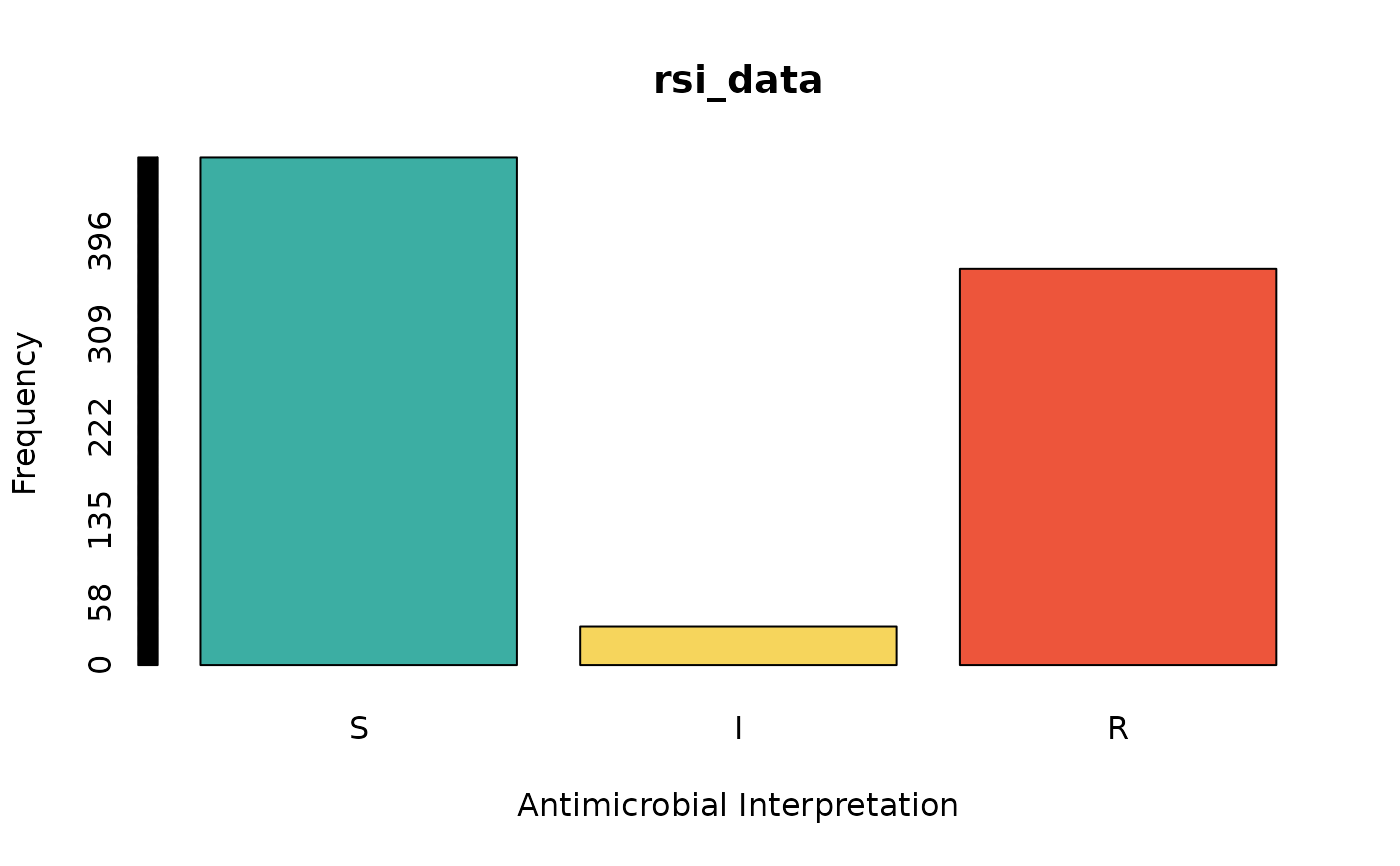 # the dplyr way
if (require("dplyr")) {
example_isolates %>%
mutate_at(vars(PEN:RIF), as.rsi)
# same:
example_isolates %>%
as.rsi(PEN:RIF)
# fastest way to transform all columns with already valid AMR results to class `rsi`:
example_isolates %>%
mutate_if(is.rsi.eligible, as.rsi)
# since dplyr 1.0.0, this can also be:
# example_isolates %>%
# mutate(across(where(is.rsi.eligible), as.rsi))
}
#> # A tibble: 2,000 × 49
#> date hospit…¹ ward_…² ward_…³ ward_…⁴ age gender patie…⁵ mo
#> <date> <fct> <lgl> <lgl> <lgl> <dbl> <chr> <chr> <mo>
#> 1 2002-01-02 D FALSE TRUE FALSE 65 F A77334 B_ESCHR_COLI
#> 2 2002-01-03 D FALSE TRUE FALSE 65 F A77334 B_ESCHR_COLI
#> 3 2002-01-07 B TRUE FALSE FALSE 45 F 067927 B_STPHY_EPDR
#> 4 2002-01-07 B TRUE FALSE FALSE 45 F 067927 B_STPHY_EPDR
#> 5 2002-01-13 B TRUE FALSE FALSE 45 F 067927 B_STPHY_EPDR
#> 6 2002-01-13 B TRUE FALSE FALSE 45 F 067927 B_STPHY_EPDR
#> 7 2002-01-14 D FALSE TRUE FALSE 78 M 462729 B_STPHY_AURS
#> 8 2002-01-14 D FALSE TRUE FALSE 78 M 462729 B_STPHY_AURS
#> 9 2002-01-16 B TRUE FALSE FALSE 45 F 067927 B_STPHY_EPDR
#> 10 2002-01-17 B TRUE FALSE FALSE 79 F 858515 B_STPHY_EPDR
#> # … with 1,990 more rows, 40 more variables: PEN <rsi>, OXA <rsi>, FLC <rsi>,
#> # AMX <rsi>, AMC <rsi>, AMP <rsi>, TZP <rsi>, CZO <rsi>, FEP <rsi>,
#> # CXM <rsi>, FOX <rsi>, CTX <rsi>, CAZ <rsi>, CRO <rsi>, GEN <rsi>,
#> # TOB <rsi>, AMK <rsi>, KAN <rsi>, TMP <rsi>, SXT <rsi>, NIT <rsi>,
#> # FOS <rsi>, LNZ <rsi>, CIP <rsi>, MFX <rsi>, VAN <rsi>, TEC <rsi>,
#> # TCY <rsi>, TGC <rsi>, DOX <rsi>, ERY <rsi>, CLI <rsi>, AZM <rsi>,
#> # IPM <rsi>, MEM <rsi>, MTR <rsi>, CHL <rsi>, COL <rsi>, MUP <rsi>, …
#> # ℹ Use `print(n = ...)` to see more rows, and `colnames()` to see all variable names
# }
# the dplyr way
if (require("dplyr")) {
example_isolates %>%
mutate_at(vars(PEN:RIF), as.rsi)
# same:
example_isolates %>%
as.rsi(PEN:RIF)
# fastest way to transform all columns with already valid AMR results to class `rsi`:
example_isolates %>%
mutate_if(is.rsi.eligible, as.rsi)
# since dplyr 1.0.0, this can also be:
# example_isolates %>%
# mutate(across(where(is.rsi.eligible), as.rsi))
}
#> # A tibble: 2,000 × 49
#> date hospit…¹ ward_…² ward_…³ ward_…⁴ age gender patie…⁵ mo
#> <date> <fct> <lgl> <lgl> <lgl> <dbl> <chr> <chr> <mo>
#> 1 2002-01-02 D FALSE TRUE FALSE 65 F A77334 B_ESCHR_COLI
#> 2 2002-01-03 D FALSE TRUE FALSE 65 F A77334 B_ESCHR_COLI
#> 3 2002-01-07 B TRUE FALSE FALSE 45 F 067927 B_STPHY_EPDR
#> 4 2002-01-07 B TRUE FALSE FALSE 45 F 067927 B_STPHY_EPDR
#> 5 2002-01-13 B TRUE FALSE FALSE 45 F 067927 B_STPHY_EPDR
#> 6 2002-01-13 B TRUE FALSE FALSE 45 F 067927 B_STPHY_EPDR
#> 7 2002-01-14 D FALSE TRUE FALSE 78 M 462729 B_STPHY_AURS
#> 8 2002-01-14 D FALSE TRUE FALSE 78 M 462729 B_STPHY_AURS
#> 9 2002-01-16 B TRUE FALSE FALSE 45 F 067927 B_STPHY_EPDR
#> 10 2002-01-17 B TRUE FALSE FALSE 79 F 858515 B_STPHY_EPDR
#> # … with 1,990 more rows, 40 more variables: PEN <rsi>, OXA <rsi>, FLC <rsi>,
#> # AMX <rsi>, AMC <rsi>, AMP <rsi>, TZP <rsi>, CZO <rsi>, FEP <rsi>,
#> # CXM <rsi>, FOX <rsi>, CTX <rsi>, CAZ <rsi>, CRO <rsi>, GEN <rsi>,
#> # TOB <rsi>, AMK <rsi>, KAN <rsi>, TMP <rsi>, SXT <rsi>, NIT <rsi>,
#> # FOS <rsi>, LNZ <rsi>, CIP <rsi>, MFX <rsi>, VAN <rsi>, TEC <rsi>,
#> # TCY <rsi>, TGC <rsi>, DOX <rsi>, ERY <rsi>, CLI <rsi>, AZM <rsi>,
#> # IPM <rsi>, MEM <rsi>, MTR <rsi>, CHL <rsi>, COL <rsi>, MUP <rsi>, …
#> # ℹ Use `print(n = ...)` to see more rows, and `colnames()` to see all variable names
# }