How to predict antimicrobial resistance
Matthijs S. Berends
23 February 2019
resistance_predict.RmdNeeded R packages
As with many uses in R, we need some additional packages for AMR analysis. Our package works closely together with the tidyverse packages dplyr and ggplot2 by Dr Hadley Wickham. The tidyverse tremendously improves the way we conduct data science - it allows for a very natural way of writing syntaxes and creating beautiful plots in R.
Our AMR package depends on these packages and even extends their use and functions.
Prediction analysis
Our package contains a function resistance_predict(), which takes the same input as functions for other AMR analysis. Based on a date column, it calculates cases per year and uses a regression model to predict antimicrobial resistance.
It is basically as easy as:
# resistance prediction of piperacillin/tazobactam (pita):
resistance_predict(tbl = septic_patients, col_date = "date", col_ab = "pita")
# or:
septic_patients %>%
resistance_predict(col_ab = "pita")
# to bind it to object 'predict_pita' for example:
predict_pita <- septic_patients %>%
resistance_predict(col_ab = "pita")The function will look for a date column itself if col_date is not set.
When running any of these commands, a summary of the regression model will be printed unless using resistance_predict(..., info = FALSE).
#> NOTE: Using column `date` as input for `col_date`.
#>
#> Logistic regression model (logit) with binomial distribution
#> ------------------------------------------------------------
#>
#> Call:
#> glm(formula = df_matrix ~ year, family = binomial)
#>
#> Deviance Residuals:
#> Min 1Q Median 3Q Max
#> -2.9224 -1.3120 0.0170 0.7586 3.1932
#>
#> Coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -222.92857 45.93922 -4.853 1.22e-06 ***
#> year 0.10994 0.02284 4.814 1.48e-06 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for binomial family taken to be 1)
#>
#> Null deviance: 59.794 on 14 degrees of freedom
#> Residual deviance: 35.191 on 13 degrees of freedom
#> AIC: 93.464
#>
#> Number of Fisher Scoring iterations: 4This text is only a printed summary - the actual result (output) of the function is a data.frame containing for each year: the number of observations, the actual observed resistance, the estimated resistance and the standard error below and above the estimation:
predict_pita
#> year value se_min se_max observations observed estimated
#> 1 2003 0.06250000 NA NA 32 0.06250000 0.06177594
#> 2 2004 0.08536585 NA NA 82 0.08536585 0.06846343
#> 3 2005 0.10000000 NA NA 60 0.10000000 0.07581637
#> 4 2006 0.05084746 NA NA 59 0.05084746 0.08388789
#> 5 2007 0.12121212 NA NA 66 0.12121212 0.09273250
#> 6 2008 0.04166667 NA NA 72 0.04166667 0.10240539
#> 7 2009 0.01639344 NA NA 61 0.01639344 0.11296163
#> 8 2010 0.09433962 NA NA 53 0.09433962 0.12445516
#> 9 2011 0.18279570 NA NA 93 0.18279570 0.13693759
#> 10 2012 0.30769231 NA NA 65 0.30769231 0.15045682
#> 11 2013 0.08620690 NA NA 58 0.08620690 0.16505550
#> 12 2014 0.15254237 NA NA 59 0.15254237 0.18076926
#> 13 2015 0.27272727 NA NA 55 0.27272727 0.19762493
#> 14 2016 0.25000000 NA NA 84 0.25000000 0.21563859
#> 15 2017 0.16279070 NA NA 86 0.16279070 0.23481370
#> 16 2018 0.25513926 0.2228376 0.2874409 NA NA 0.25513926
#> 17 2019 0.27658825 0.2386811 0.3144954 NA NA 0.27658825
#> 18 2020 0.29911630 0.2551715 0.3430611 NA NA 0.29911630
#> 19 2021 0.32266085 0.2723340 0.3729877 NA NA 0.32266085
#> 20 2022 0.34714076 0.2901847 0.4040968 NA NA 0.34714076
#> 21 2023 0.37245666 0.3087318 0.4361815 NA NA 0.37245666
#> 22 2024 0.39849187 0.3279750 0.4690088 NA NA 0.39849187
#> 23 2025 0.42511415 0.3479042 0.5023241 NA NA 0.42511415
#> 24 2026 0.45217796 0.3684992 0.5358568 NA NA 0.45217796
#> 25 2027 0.47952757 0.3897276 0.5693275 NA NA 0.47952757
#> 26 2028 0.50700045 0.4115444 0.6024565 NA NA 0.50700045
#> 27 2029 0.53443111 0.4338908 0.6349714 NA NA 0.53443111The function plot is available in base R, and can be extended by other packages to depend the output based on the type of input. We extended its function to cope with resistance predictions:
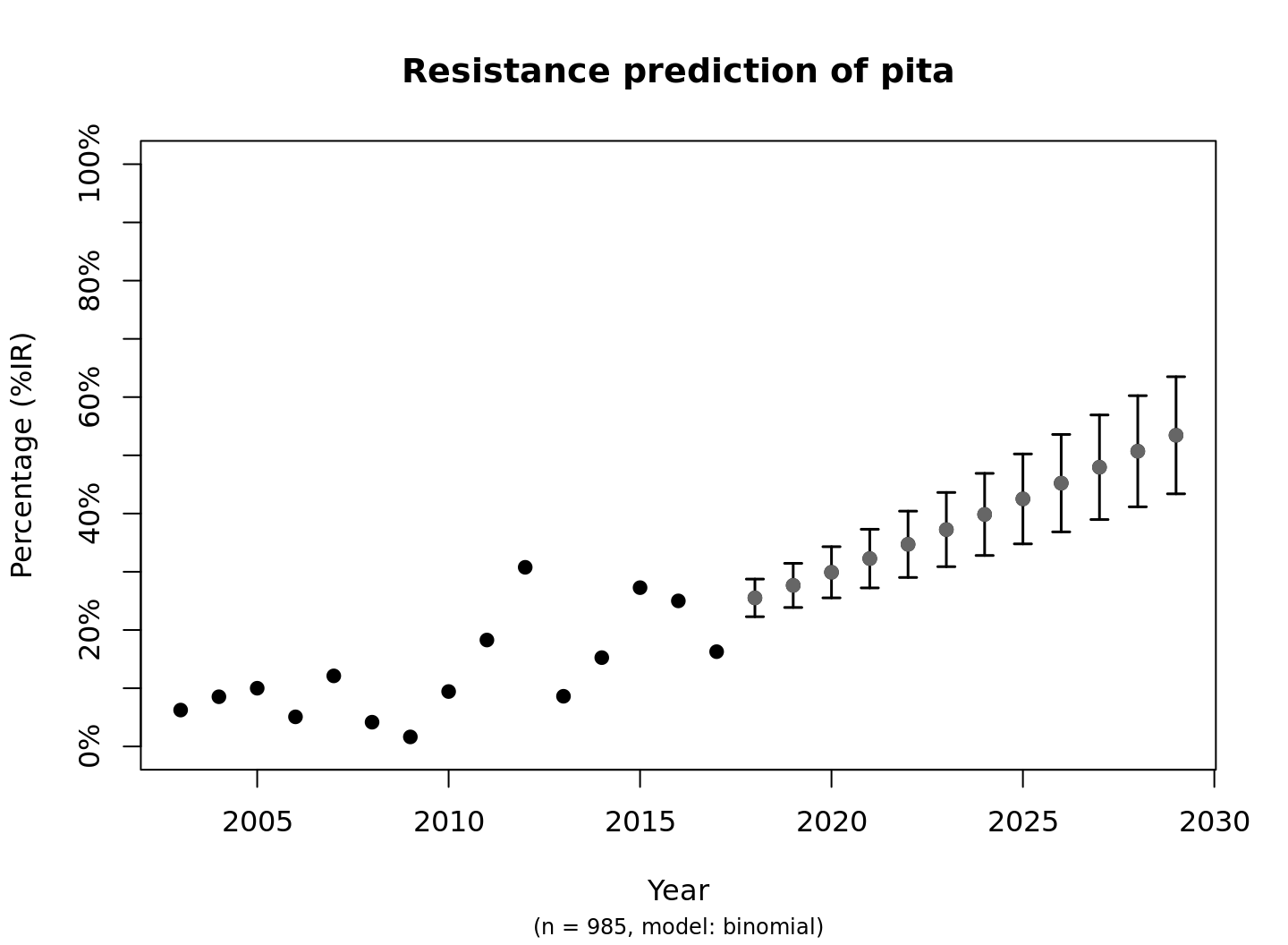
This is the fastest way to plot the result. It automatically adds the right axes, error bars, titles, number of available observations and type of model.
We also support the ggplot2 package with our custom function ggplot_rsi_predict() to create more appealing plots:
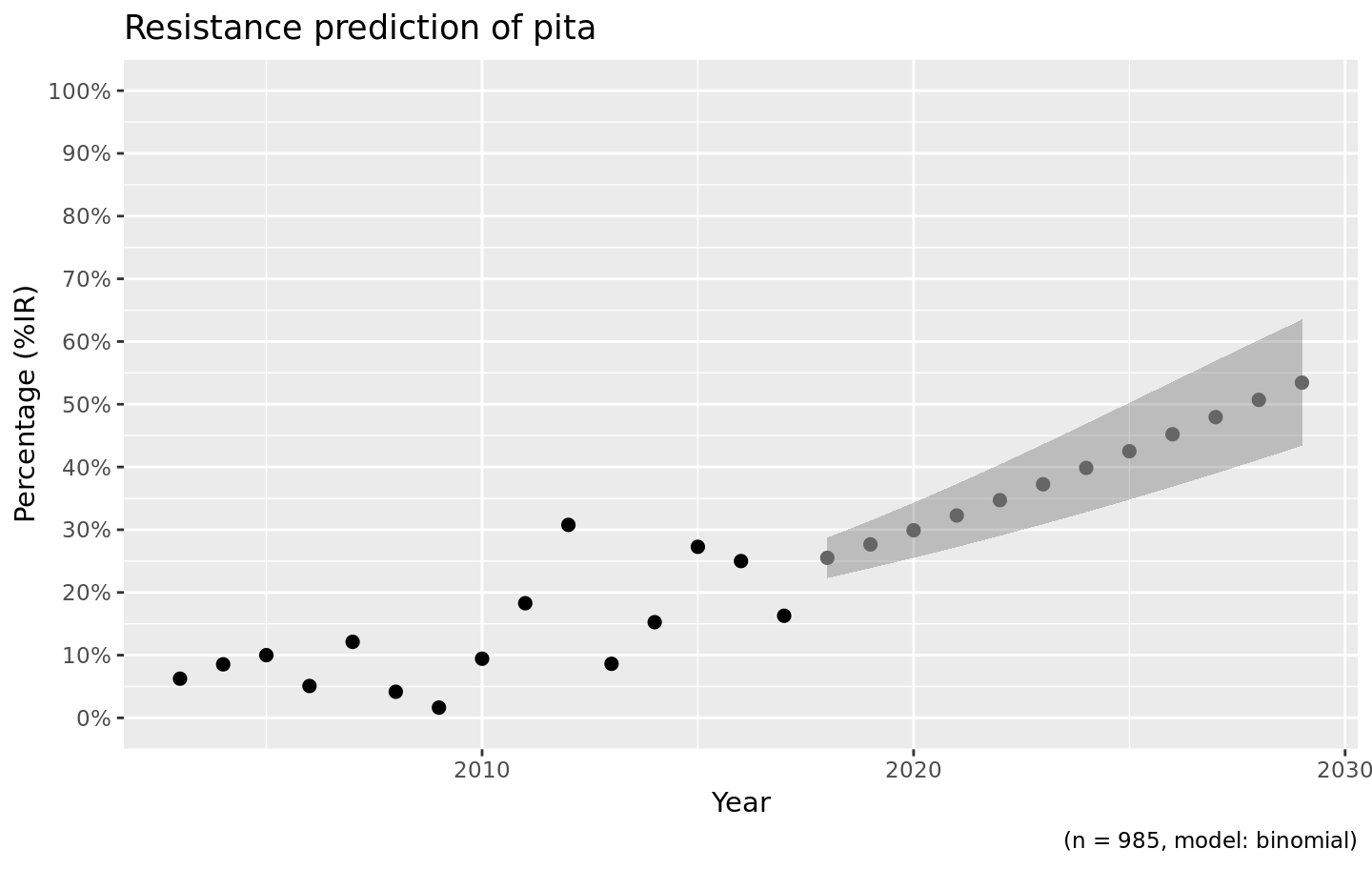
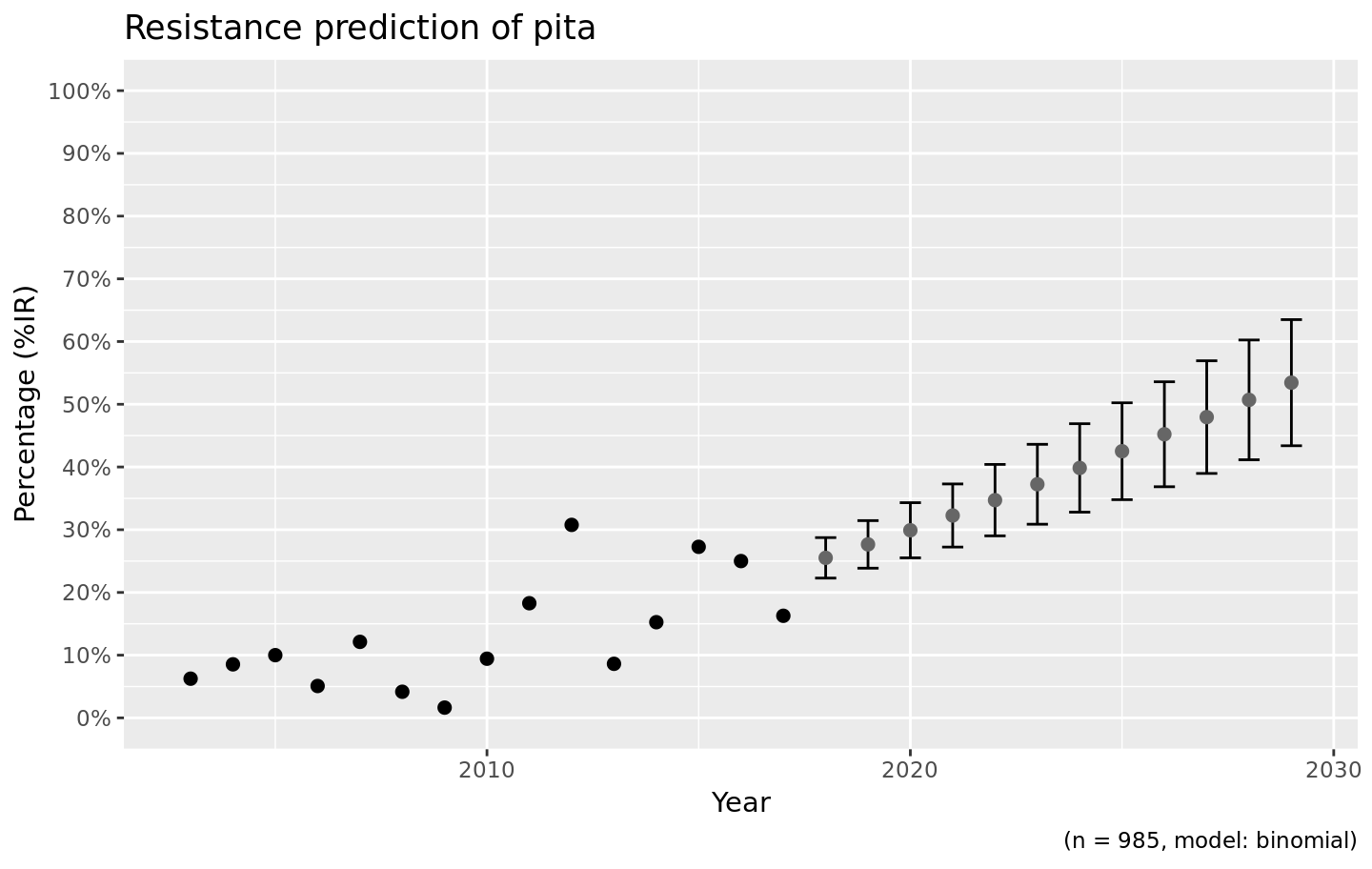
Choosing the right model
Resistance is not easily predicted; if we look at vancomycin resistance in Gram positives, the spread (i.e. standard error) is enormous:
septic_patients %>%
filter(mo_gramstain(mo) == "Gram positive") %>%
resistance_predict(col_ab = "vanc", year_min = 2010, info = FALSE) %>%
ggplot_rsi_predict()
#> [1] "is any"
#> [1] "is any"
#> NOTE: Using column `date` as input for `col_date`.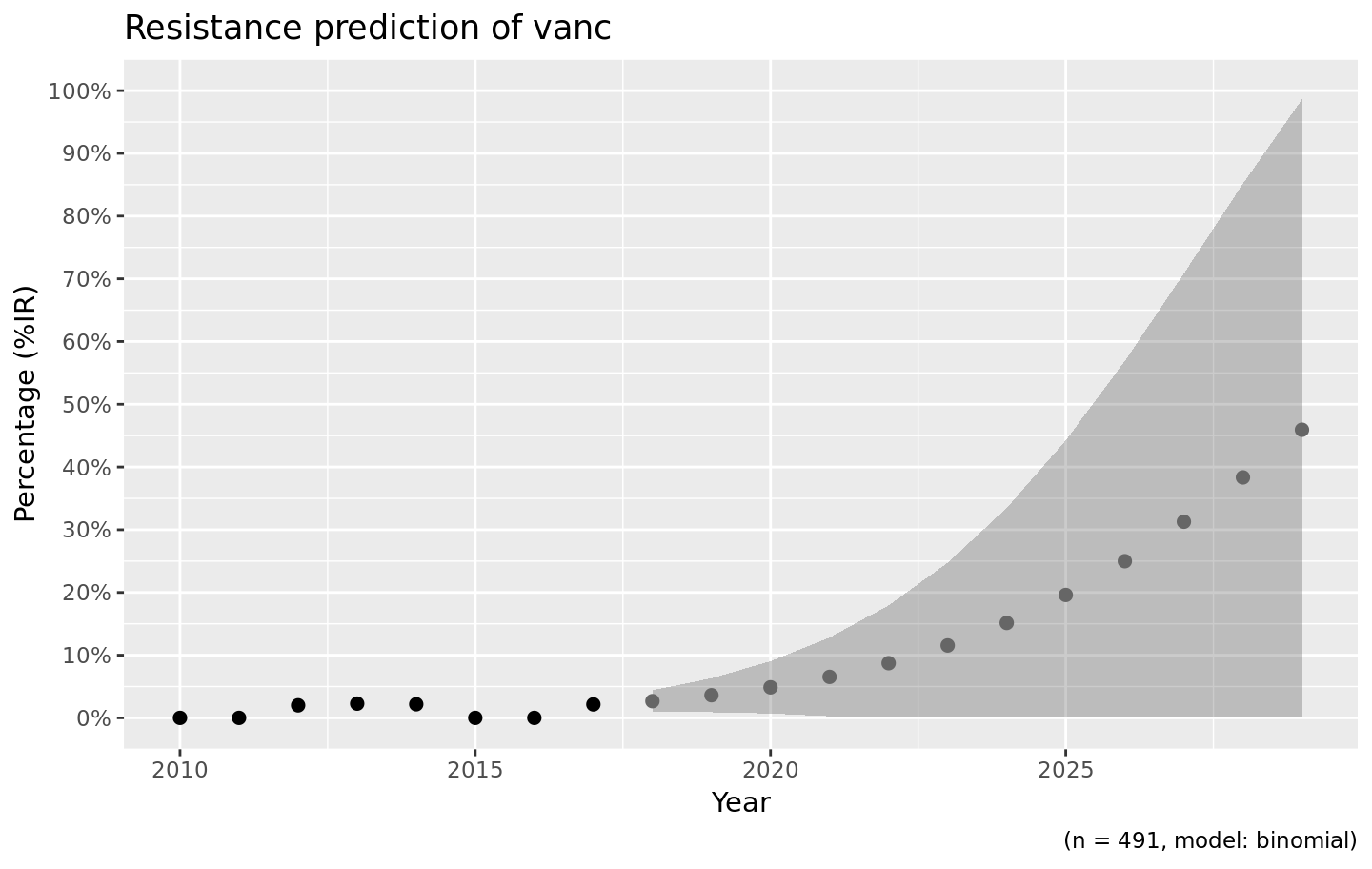
Vancomycin resistance could be 100% in ten years, but might also stay around 0%.
You can define the model with the model parameter. The default model is a generalised linear regression model using a binomial distribution, assuming that a period of zero resistance was followed by a period of increasing resistance leading slowly to more and more resistance.
Valid values are:
| Input values | Function used by R | Type of model |
|---|---|---|
"binomial" or "binom" or "logit"
|
glm(..., family = binomial) |
Generalised linear model with binomial distribution |
"loglin" or "poisson"
|
glm(..., family = poisson) |
Generalised linear model with poisson distribution |
"lin" or "linear"
|
lm() |
Linear model |
For the vancomycin resistance in Gram positive bacteria, a linear model might be more appropriate since no (left half of a) binomial distribution is to be expected based on the observed years:
septic_patients %>%
filter(mo_gramstain(mo) == "Gram positive") %>%
resistance_predict(col_ab = "vanc", year_min = 2010, info = FALSE, model = "linear") %>%
ggplot_rsi_predict()
#> [1] "is any"
#> [1] "is any"
#> NOTE: Using column `date` as input for `col_date`.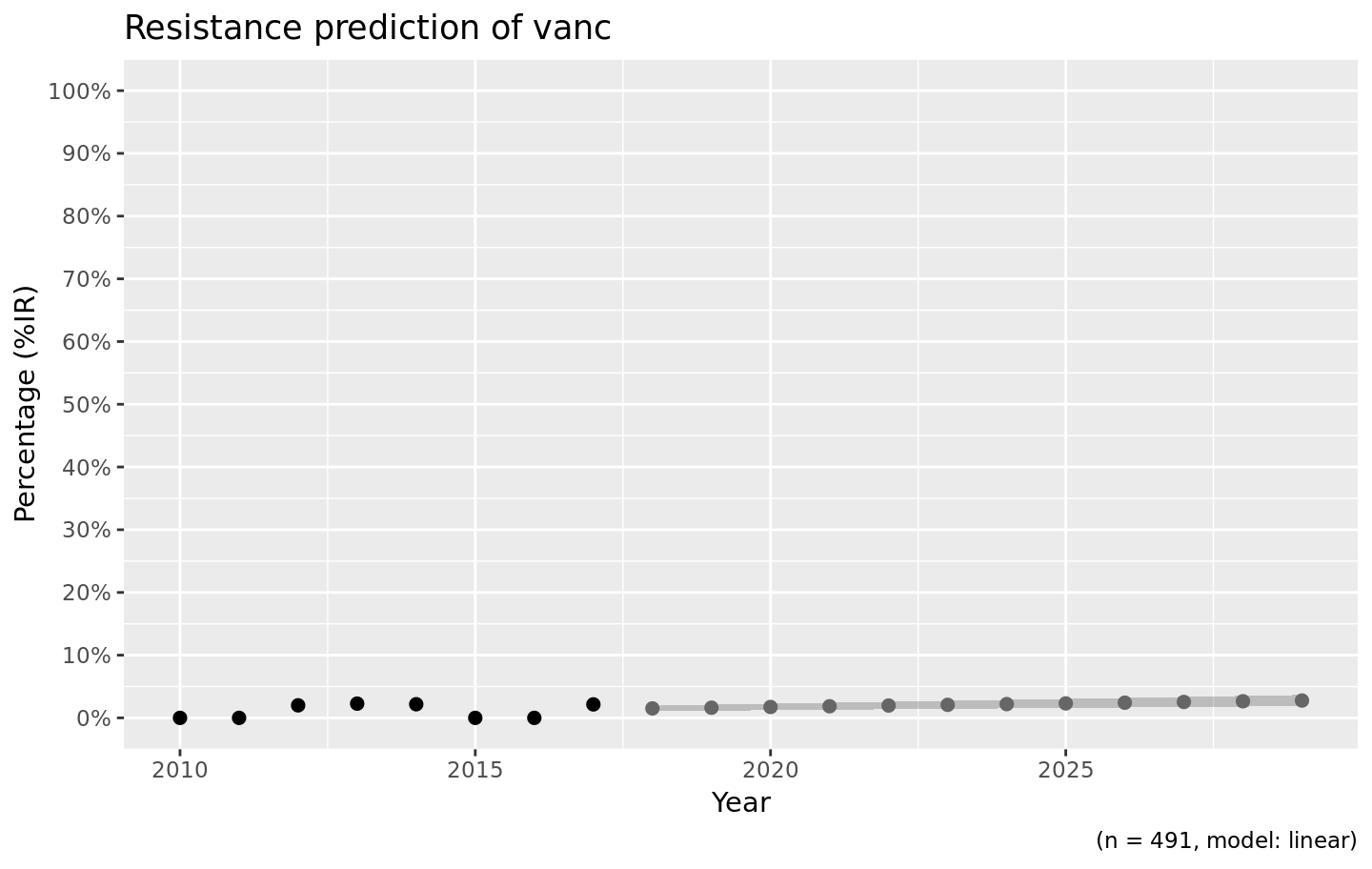
This seems more likely, doesn’t it?
The model itself is also available from the object, as an attribute: