AMR
An R package to simplify the analysis and prediction of Antimicrobial Resistance (AMR) and work with antibiotic properties by using evidence-based methods.
This R package was created for academic research by PhD students of the Faculty of Medical Sciences of the University of Groningen and the Medical Microbiology & Infection Prevention (MMBI) department of the University Medical Center Groningen (UMCG).
▶️ Download it with install.packages("AMR") or see below for other possibilities.
Authors
Matthijs S. Berends1,2,a,
Christian F. Luz1,a,
Erwin E.A. Hassing2,
Corinna Glasner1,b,
Alex W. Friedrich1,b,
Bhanu Sinha1,b
1 Department of Medical Microbiology, University of Groningen, University Medical Center Groningen, Groningen, the Netherlands - rug.nl umcg.nl
2 Certe Medical Diagnostics & Advice, Groningen, the Netherlands - certe.nl
a R package author and thesis dissertant
b Thesis advisor
Contents
Why this package?
This R package was intended to make microbial epidemiology easier. Most functions contain extensive help pages to get started.
With AMR you can:
- Calculate the resistance (and even co-resistance) of microbial isolates with the
portion_R,portion_IR,portion_I,portion_SIandportion_Sfunctions, that can also be used with thedplyrpackage (e.g. in conjunction withsummarise) - Plot AMR results with
geom_rsi, a function made for theggplot2package - Predict antimicrobial resistance for the nextcoming years using logistic regression models with the
resistance_predictfunction - Apply EUCAST rules to isolates with the
EUCAST_rulesfunction - Identify first isolates of every patient using guidelines from the CLSI (Clinical and Laboratory Standards Institute) with the
first_isolatefunction- You can also identify first weighted isolates of every patient, an adjusted version of the CLSI guideline. This takes into account key antibiotics of every strain and compares them. The following 12 antibiotics will be used as key antibiotics at default:
- Universal: amoxicillin, amoxicillin/clavlanic acid, cefuroxime, piperacillin/tazobactam, ciprofloxacin, trimethoprim/sulfamethoxazole
- Specific for Gram-positives: vancomycin, teicoplanin, tetracycline, erythromycin, oxacillin, rifampicin
- Specific for Gram-negatives: gentamicin, tobramycin, colistin, cefotaxime, ceftazidime, meropenem
- You can also identify first weighted isolates of every patient, an adjusted version of the CLSI guideline. This takes into account key antibiotics of every strain and compares them. The following 12 antibiotics will be used as key antibiotics at default:
- Categorise Staphylococci into Coagulase Negative Staphylococci (CoNS) and Coagulase Positve Staphylococci (CoPS) according to Karsten Becker et al.
- Categorise Streptococci into Lancefield groups
- Get antimicrobial ATC properties from the WHO Collaborating Centre for Drug Statistics Methodology (WHOCC), to be able to:
- Translate antibiotic codes (like AMOX), official names (like amoxicillin) and even trade names (like Amoxil or Trimox) to an ATC code (like J01CA04) and vice versa with the
abnamefunction - Get the latest antibiotic properties like hierarchic groups and defined daily dose (DDD) with units and administration form from the WHOCC website with the
atc_propertyfunction
- Translate antibiotic codes (like AMOX), official names (like amoxicillin) and even trade names (like Amoxil or Trimox) to an ATC code (like J01CA04) and vice versa with the
- Conduct descriptive statistics: calculate kurtosis, skewness and create frequency tables
And it contains:
- A recent data set with ~2500 human pathogenic microorganisms, including family, genus, species, gram stain and aerobic/anaerobic
- A recent data set with all antibiotics as defined by the WHOCC, including ATC code, official name and DDD's
- An example data set
septic_patients, consisting of 2000 blood culture isolates from anonymised septic patients between 2001 and 2017.
With the MDRO function (abbreviation of Multi Drug Resistant Organisms), you can check your isolates for exceptional resistance with country-specific guidelines or EUCAST rules. Currently guidelines for Germany and the Netherlands are supported. Please suggest addition of your own country here: https://github.com/msberends/AMR/issues/new.
Read all changes and new functions in NEWS.md.
How to get it?
All versions of this package are published on CRAN, the official R network with a peer-reviewed submission process.
Install from CRAN
(Note: Downloads measured only by cran.rstudio.com, this excludes e.g. the official cran.r-project.org)
-
Install using RStudio (recommended):
- Click on
Toolsand thenInstall Packages... - Type in
AMRand press Install
- Click on
-
install.packages("AMR")
Install from GitHub
This is the latest development version. Although it may contain bugfixes and even new functions compared to the latest released version on CRAN, it is also subject to change and may be unstable or behave unexpectedly. Always consider this a beta version.
install.packages("devtools")
devtools::install_github("msberends/AMR")
Install from Zenodo
This package was also published on Zenodo: https://doi.org/10.5281/zenodo.1305355
How to use it?
# Call it with:
library(AMR)
# For a list of functions:
help(package = "AMR")
New classes
This package contains two new S3 classes: mic for MIC values (e.g. from Vitek or Phoenix) and rsi for antimicrobial drug interpretations (i.e. S, I and R). Both are actually ordered factors under the hood (an MIC of 2 being higher than <=1 but lower than >=32, and for class rsi factors are ordered as S < I < R).
Both classes have extensions for existing generic functions like print, summary and plot.
These functions also try to coerce valid values.
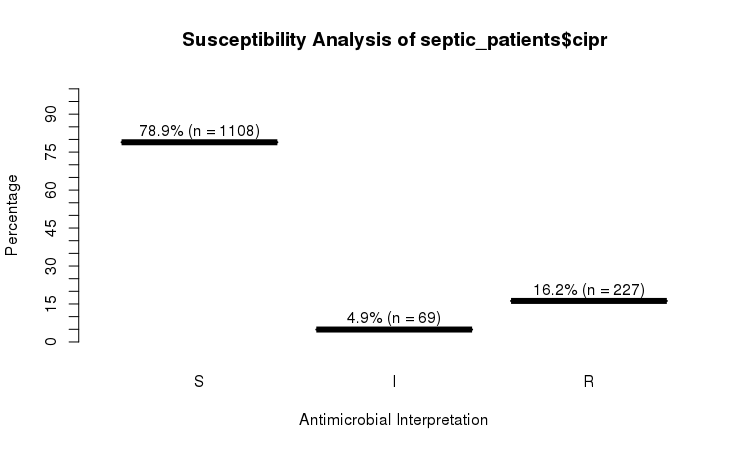
RSI
The septic_patients data set comes with antimicrobial results of more than 40 different drugs. For example, columns amox and cipr contain results of amoxicillin and ciprofloxacin, respectively.
summary(septic_patients[, c("amox", "cipr")])
# amox cipr
# Mode :rsi Mode :rsi
# <NA> :1002 <NA> :596
# Sum S :336 Sum S :1108
# Sum IR:662 Sum IR:296
# -Sum R:659 -Sum R:227
# -Sum I:3 -Sum I:69
You can use the plot function from base R:
plot(septic_patients$cipr)
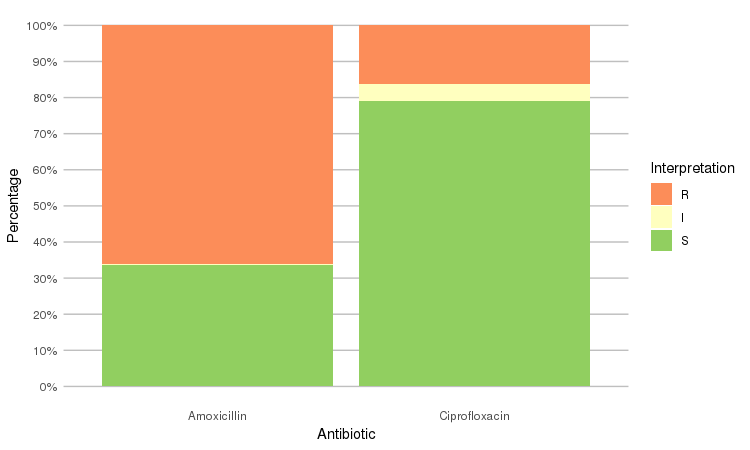
Or use the ggplot2 and dplyr packages to create more appealing plots:
library(dplyr)
library(ggplot2)
septic_patients %>%
select(amox, cipr) %>%
ggplot_rsi()
septic_patients %>%
select(amox, cipr) %>%
ggplot_rsi(x = "Interpretation", facet = "Antibiotic")
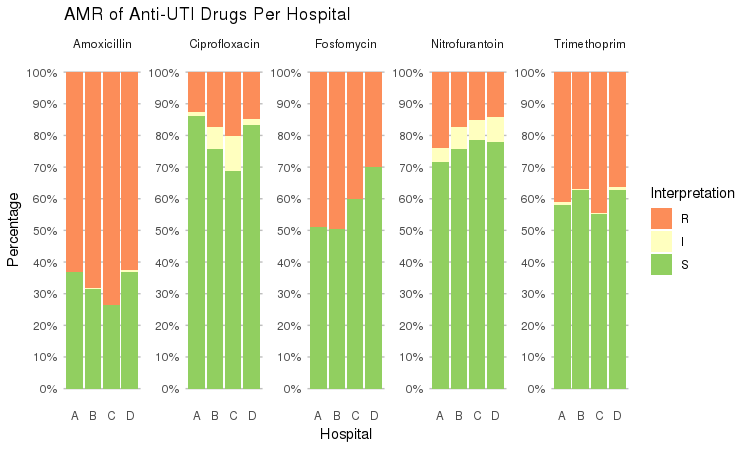
It also supports grouping variables. Let's say we want to compare resistance of drugs against Urine Tract Infections (UTI) between hospitals A to D (variable hospital_id):
septic_patients %>%
select(hospital_id, amox, nitr, fosf, trim, cipr) %>%
group_by(hospital_id) %>%
ggplot_rsi(x = "hospital_id",
facet = "Antibiotic",
nrow = 1) +
labs(title = "AMR of Anti-UTI Drugs Per Hospital",
x = "Hospital")
You could use this to group on anything in your plots: Gram stain, age (group), genus, geographic location, et cetera.
MIC
# Transform values to new class
mic_data <- as.mic(c(">=32", "1.0", "8", "<=0.128", "8", "16", "16"))
summary(mic_data)
# Mode:mic
# <NA>:0
# Min.:<=0.128
# Max.:>=32
plot(mic_data)
Overwrite/force resistance based on EUCAST rules
This is also called interpretive reading.
before <- data.frame(bactid = c("STAAUR", # Staphylococcus aureus
"ENCFAE", # Enterococcus faecalis
"ESCCOL", # Escherichia coli
"KLEPNE", # Klebsiella pneumoniae
"PSEAER"), # Pseudomonas aeruginosa
vanc = "-", # Vancomycin
amox = "-", # Amoxicillin
coli = "-", # Colistin
cfta = "-", # Ceftazidime
cfur = "-", # Cefuroxime
stringsAsFactors = FALSE)
before
# bactid vanc amox coli cfta cfur
# 1 STAAUR - - - - -
# 2 ENCFAE - - - - -
# 3 ESCCOL - - - - -
# 4 KLEPNE - - - - -
# 5 PSEAER - - - - -
# Now apply those rules; just need a column with bacteria ID's and antibiotic results:
after <- EUCAST_rules(before)
after
# bactid vanc amox coli cfta cfur
# 1 STAAUR - - R R -
# 2 ENCFAE - - R R R
# 3 ESCCOL R - - - -
# 4 KLEPNE R R - - -
# 5 PSEAER R R - - R
Bacteria ID's can be retrieved with the guess_bactid function. It uses any type of info about a microorganism as input. For example, all these will return value STAAUR, the ID of S. aureus:
guess_bactid("stau")
guess_bactid("STAU")
guess_bactid("staaur")
guess_bactid("S. aureus")
guess_bactid("S aureus")
guess_bactid("Staphylococcus aureus")
guess_bactid("MRSA") # Methicillin Resistant S. aureus
guess_bactid("VISA") # Vancomycin Intermediate S. aureus
guess_bactid("VRSA") # Vancomycin Resistant S. aureus
Other (microbial) epidemiological functions
# G-test to replace Chi squared test
g.test(...)
# Determine key antibiotic based on bacteria ID
key_antibiotics(...)
# Selection of first isolates of any patient
first_isolate(...)
# Calculate resistance levels of antibiotics, can be used with `summarise` (dplyr)
rsi(...)
# Predict resistance levels of antibiotics
rsi_predict(...)
# Get name of antibiotic by ATC code
abname(...)
abname("J01CR02", from = "atc", to = "umcg") # "AMCL"
Frequency tables
Base R lacks a simple function to create frequency tables. We created such a function that works with almost all data types: freq (or frequency_tbl). It can be used in two ways:
# Like base R:
freq(mydata$myvariable)
# And like tidyverse:
mydata %>% freq(myvariable)
Factors sort on item by default:
septic_patients %>% freq(hospital_id)
# Frequency table of `hospital_id`
# Class: factor
# Length: 2000 (of which NA: 0 = 0.0%)
# Unique: 4
#
# Item Count Percent Cum. Count Cum. Percent (Factor Level)
# --- ----- ------ -------- ----------- ------------- ---------------
# 1 A 319 16.0% 319 16.0% 1
# 2 B 661 33.1% 980 49.0% 2
# 3 C 256 12.8% 1236 61.8% 3
# 4 D 764 38.2% 2000 100.0% 4
This can be changed with the sort.count parameter:
septic_patients %>% freq(hospital_id, sort.count = TRUE)
# Frequency table of `hospital_id`
# Class: factor
# Length: 2000 (of which NA: 0 = 0.0%)
# Unique: 4
#
# Item Count Percent Cum. Count Cum. Percent (Factor Level)
# --- ----- ------ -------- ----------- ------------- ---------------
# 1 D 764 38.2% 764 38.2% 4
# 2 B 661 33.1% 1425 71.2% 2
# 3 A 319 16.0% 1744 87.2% 1
# 4 C 256 12.8% 2000 100.0% 3
All other types, like numbers, characters and dates, sort on count by default:
septic_patients %>% freq(date)
# Frequency table of `date`
# Class: Date
# Length: 2000 (of which NA: 0 = 0.0%)
# Unique: 1151
#
# Oldest: 2 January 2002
# Newest: 28 December 2017 (+5839)
# Median: 7 Augustus 2009 (~48%)
#
# Item Count Percent Cum. Count Cum. Percent
# --- ----------- ------ -------- ----------- -------------
# 1 2016-05-21 10 0.5% 10 0.5%
# 2 2004-11-15 8 0.4% 18 0.9%
# 3 2013-07-29 8 0.4% 26 1.3%
# 4 2017-06-12 8 0.4% 34 1.7%
# 5 2015-11-19 7 0.4% 41 2.1%
# 6 2005-12-22 6 0.3% 47 2.4%
# 7 2015-10-12 6 0.3% 53 2.6%
# 8 2002-05-16 5 0.2% 58 2.9%
# 9 2004-02-02 5 0.2% 63 3.1%
# 10 2004-02-18 5 0.2% 68 3.4%
# 11 2005-08-16 5 0.2% 73 3.6%
# 12 2005-09-01 5 0.2% 78 3.9%
# 13 2006-06-29 5 0.2% 83 4.2%
# 14 2007-08-10 5 0.2% 88 4.4%
# 15 2008-08-29 5 0.2% 93 4.7%
# [ reached getOption("max.print.freq") -- omitted 1136 entries, n = 1907 (95.3%) ]
For numeric values, some extra descriptive statistics will be calculated:
freq(runif(n = 10, min = 1, max = 5))
# Frequency table
# Class: numeric
# Length: 10 (of which NA: 0 = 0.0%)
# Unique: 10
#
# Mean: 3.4
# Std. dev.: 1.3 (CV: 0.38, MAD: 1.3)
# Five-Num: 1.6 | 2.0 | 3.9 | 4.7 | 4.8 (IQR: 2.7, CQV: 0.4)
# Outliers: 0
#
# Item Count Percent Cum. Count Cum. Percent
# --- --------- ------ -------- ----------- -------------
# 1 1.568997 1 10.0% 1 10.0%
# 2 1.993575 1 10.0% 2 20.0%
# 3 2.022348 1 10.0% 3 30.0%
# 4 2.236038 1 10.0% 4 40.0%
# 5 3.579828 1 10.0% 5 50.0%
# 6 4.178081 1 10.0% 6 60.0%
# 7 4.394818 1 10.0% 7 70.0%
# 8 4.689871 1 10.0% 8 80.0%
# 9 4.698626 1 10.0% 9 90.0%
# 10 4.751488 1 10.0% 10 100.0%
#
# Warning message:
# All observations are unique.
Learn more about this function with:
?freq
Data sets included in package
Datasets to work with antibiotics and bacteria properties.
# Dataset with 2000 random blood culture isolates from anonymised
# septic patients between 2001 and 2017 in 5 Dutch hospitals
septic_patients # A tibble: 2,000 x 49
# Dataset with ATC antibiotics codes, official names, trade names
# and DDD's (oral and parenteral)
antibiotics # A tibble: 420 x 18
# Dataset with bacteria codes and properties like gram stain and
# aerobic/anaerobic
microorganisms # A tibble: 2,453 x 12
Copyright
This R package is licensed under the GNU General Public License (GPL) v2.0. In a nutshell, this means that this package:
-
May be used for commercial purposes
-
May be used for private purposes
-
May not be used for patent purposes
-
May be modified, although:
- Modifications must be released under the same license when distributing the package
- Changes made to the code must be documented
-
May be distributed, although:
- Source code must be made available when the package is distributed
- A copy of the license and copyright notice must be included with the package.
-
Comes with a LIMITATION of liability
-
Comes with NO warranty